Cross-Disorder Genomewide Analysis of Schizophrenia, Bipolar Disorder, and Depression
Abstract
Objective:
Family and twin studies indicate substantial overlap of genetic influences on psychotic and mood disorders. Linkage and candidate gene studies have also suggested overlap across schizophrenia, bipolar disorder, and major depressive disorder. The purpose of this study was to apply genomewide association study (GWAS) analysis to address the specificity of genetic effects on these disorders.
Method:
The authors combined GWAS data from three large effectiveness studies of schizophrenia (CATIE, genotyped: N=741), bipolar disorder (STEP-BD, geno-typed: N=1,575), and major depressive disorder (STAR*D, genotyped: N=1,938) as well as from psychiatrically screened control subjects (NIMH-Genetics Repository: N=1,204). A two-stage analytic procedure involving an omnibus test of allele frequency differences among case and control groups was applied, followed by a model selection step to identify the best-fitting model of allelic effects across disorders.
Results:
The strongest result was seen for a single nucleotide polymorphism near the adrenomedullin (ADM) gene (rs6484218), with the best-fitting model indicating that the effect was specific to bipolar II disorder. Findings also revealed evidence suggesting that several genes may have effects that transcend clinical diagnostic boundaries, including variants in NPAS3 that showed pleiotropic effects across schizophrenia, bipolar disorder, and major depressive disorder.
Conclusions:
This study provides the first genomewide significant evidence implicating variants near the ADM gene on chromosome 11p15 in psychopathology, with effects that appear to be specific to bipolar II disorder. Although genomewide signifi-cant evidence of cross-disorder effects was not detected, the results provide evidence that there are both pleiotropic and disorder-specific effects on major mental illness and illustrate an approach to dissecting the genetic basis of mood and psychotic disorders that can inform future large-scale cross-disorder GWAS analyses.
Family and twin studies have established that schizophrenia, bipolar disorder, and major depressive disorder are familial and heritable phenotypes and that genetic factors are the most robustly validated risk factors for each disorder (1–3). However, several findings have called into question whether these disorders are etiologically distinct. First, several key clinical features, including psychosis, neurocognitive impairment, and suicidality, may be observed in all three disorders. Second, genetic epidemio-logic studies have documented that schizophrenia, bipolar disorder, and major depressive disorder share familial and genetic determinants. Family studies have shown familial coaggregation for schizophrenia and bipolar disorder (4–6) as well as bipolar disorder and major depressive disorder (1). In a population-based study of more than 2 million families, Lichtenstein et al. (7) demonstrated increased risks of schizophrenia among relatives of bipolar disorder probands and increased risks of bipolar disorder among relatives of schizophrenia probands. Comorbidity between the disorders was mainly attributable to overlapping genetic influences. Twin studies have similarly documented substantial shared genetic variance between psychotic disorders and bipolar disorder (8) and between bipolar disorder and major depressive disorder (9).
Although family and twin studies can estimate the shared heritability across disorders, they cannot identify the genetic loci contributing to this overlap. To date, evidence implicating specific chromosomal regions and genes in the shared liability to psychotic and mood disorders has largely been limited to linkage and candidate gene association studies. Some of the regions with the strongest linkage evidence for schizophrenia are also among regions most strongly linked to bipolar disorder (10–12), although simulations suggest that such overlap could easily occur by chance (7). Several chromosomal microdeletions have also been associated with both mood and psychotic disorders. The balanced translocation ([1, 11][q42;q14.3]) that disrupts the DISC1 gene was first identified because of its cosegregation with a broad phenotype comprising schizophrenia, bipolar disorder, and recurrent major depressive disorder (13). The 22q11 microdeletion responsible for velocardiofacial syndrome also appears to confer increased risk of both psychotic and mood disorders (14, 15). Candidate gene studies have also found association between specific genes and both psychotic and mood disorder phenotypes (11, 16, 17), although results have been inconsistent (18, 19).
Genomewide association studies (GWAS), which provide a survey of common genetic variation across the genome, offer a more comprehensive method for identifying risk loci at the genotypic level. Early efforts to apply this technology to major psychiatric disorders have begun to bear fruit, with GWAS implicating several susceptibility genes for schizophrenia (20), bipolar disorder (21), and major depressive disorder (22). A recent analysis examined genewide evidence of association using data from both a bipolar disorder and schizophrenia GWAS, respectively, and found nominal evidence that several genes influence both disorders (23). Data from the International Schizophrenia Consortium demonstrated that common genetic variation (involving thousands of small-effect alleles) accounts for at least one-third of the total variation in liability to schizophrenia and that these polygenic risks are substantially shared with bipolar disorder (24). To date, however, no GWAS have been reported that examine cross-disorder analyses of the specificity of genetic influences for all three disorders. In the present study, we report the first genomewide cross-disorder analysis incorporating samples from the three largest treatment effectiveness studies of schizophrenia, bipolar disorder, and major depressive disorder, respectively. To address the issue of multiple comparisons, we utilized a novel approach that examines the patterns of cross-disorder effects in a single-model selection framework that controls the type I error risk at an experiment-wise level.
Method
Clinical Samples
Bipolar disorder (Systematic Treatment Enhancement Program for Bipolar Disorder [STEP-BD]).
STEP-BD was a national, longitudinal public health initiative designed to examine the effectiveness of treatments and their impact on the course of bipolar disorder (25). Over a 7-year period, 4,361 participants were enrolled across 20 sites and followed for up to 2 years. To maximize external validity, enrollment was offered to all eligible patients seeking outpatient treatment at one of the participating sites (26). Eligibility for STEP-BD required a consensus DSM-IV bipolar diagnosis on both the Affective Disorders Evaluation and Mini-International Neuropsychiatric Interview-PLUS semistructured interviews, as previously described (25). From the parent STEP-BD study, 2,089 individuals were enrolled in a genetic substudy.
Schizophrenia (Clinical Antipsychotic Trials of Intervention Effectiveness [CATIE]).
CATIE was a multiphase randomized controlled trial of antipsychotic medications comprising 1,460 individuals with schizophrenia followed for up to 18 months (27, 28). Final study diagnoses of DSM-IV schizophrenia were established by CATIE clinicians using the Structured Clinical Interview for DSM-IV (29), including review of all available information (encompassing psychiatric and general medical records). As detailed previously (27), exclusion criteria included diagnosis of schizoaffective disorder, mental retardation or other cognitive disorder, single psychotic episode, and history of treatment resistance or serious adverse reaction to the study treatments. As previously described (30), the genetic substudy included 738 cases.
Major depressive disorder (Sequenced Treatment Alternatives to Relieve Depression [STAR*D]).
STAR*D was a multi-site, prospective, randomized multiphase clinical trial of outpatients with nonpsychotic major depressive disorder that enrolled 4,041 participants over a 3-year period (31). Eligibility required a single or recurrent nonpsychotic major depressive episode (by DSM-IV criteria) and a score of ≥14 on the 17-item Hamilton Depression Rating Scale. Relevant exclusion criteria included history of bipolar disorder, schizophrenia, schizoaffective disorder, or psychosis not otherwise specified. In the genetic substudy, blood samples were collected from 1,953 participants.
Control (National Institute of Mental Health [NIMH] Genetics Repository).
As previously described (32), control subjects were collected by Knowledge Networks, a survey and market research company whose panel contains approximately 60,000 households representative of the U.S. population. Subjects completed an online psychiatric screen that included questions regarding demographics, ancestry, and DSM-IV criteria for a range of psychiatric disorders. Participants who reported a history of schizophrenia, psychosis, or bipolar disorder were excluded from the GWAS analyses, as previously described (32). We also excluded individuals (N=126) who met criteria for a history of major depressive episode.
Genotyping
Genotyping of STEP-BD and CATIE samples was performed using the Affymetrix GeneChip Human Mapping 500K Array Set (Affymetrix, Inc., Santa Clara, Calif.), while one-half of the STAR*D sample was genotyped with the 500K array and one-half with the Affymetrix Human Single Nucleotide Polymorphism (SNP) Array, 5.0. Genotyping of the STEP-BD and control samples was performed at the Broad Institute, as previously described (33). Quality control processing of genotypes was described by Sklar et al. (32). Genotyping of the schizophrenia sample was performed at Perlegen Sciences (Mountain View, Calif.), as reported elsewhere (30). Genotyping of the STAR*D sample was performed at Affymetrix (500K) or at the University of California, San Francisco (5.0), as described previously (34).
Quality control and harmonization of genotype data.
Additional quality control for the combined genotypic data set was performed using PLINK (35), as previously described (32).
In brief, individuals were excluded if they had overall call rates <95%, excess or insufficient heterozygosity, or apparent relatedness. We only included non-Hispanic Caucasian individuals with European ancestry based on self-reported race and ethnicity information. The PLINK nearest neighbor method (35) was used to filter out potential outliers based on the first 10 multidimensional scaling factors. SNPs were excluded if they had a call rate <98%, had a minor allele frequency <1%, were inconsistent with Hardy-Weinberg equilibrium at a p value of <1×10–6, or showed differential rates of missingness in patient and control cases (32). After quality control steps were performed, 224,395 genotyped markers were retained, with a total genotyping rate in the final sample >99%. This set of genotyped markers is smaller than that reported for the primary GWAS reports of each sample (30, 32) because of the aforementioned additional quality control steps. We used BEAGLE, Version 3.1.1, (http://www.stat.auckland.ac.nz/∼bbrowning/beagle/beagle.html) to impute missing geno-types, with HapMap (Centre d'Etude du Polymorphisme Humain from Utah population, release 23, forward strand) as the reference panel. We excluded SNPs with an imputation quality R2 score <0.8 and obtained a total of 1,574,154 SNPs for the final analysis. For each SNP, imputed dosage was then summed for each of the five phenotype groups.
Further control for confounding by population stratification was performed by calculating the first 10 quantitative ancestry indices based on the merged data set using multidimensional scaling analysis (35). To examine the effect of each of the 10 quantitative indices (C1–C10), we calculated the genomic inflation factor (λ) from a GWA analysis of genotypes using each quantitative index as the dependent variable. We plotted the λ's against each of the 10 indices (analogous to a scree plot) and found excessive values only for C1–C4. Thus, we used these four indices as covariates in our GWAS of the phenotypic groups.
Statistical Analysis
Examining the pleiotropic of genetic variants across disorders in the GWAS context requires additional attention to problems of multiple testing and type I error. One approach to the analysis is to conduct a series of pairwise comparisons of the individual disorders and their combinations. However, this would effectively entail conducting multiple GWAS analyses, each of which would require correction for multiple testing. We have taken an alternative approach that involves a single genomewide omnibus test of association across disorders followed by a model selection approach that asks which configuration of phenotypes is most likely to be associated with a given variant. The sequence of analytic steps was as follows:
We first computed a likelihood ratio statistic (the omnibus test statistic) from a multinomial logistic regression in which allele frequencies can vary for each sample (schizophrenia, bipolar I disorder, bipolar II disorder, major depressive disorder, and control) relative to a null model in which allele frequencies are the same across all groups. This 4-df test is essentially a test of whether there is any association between the variant and any of the four target disorders.
Next, for each SNP whose omnibus test resulted in p<5×10-5, we fit nine additional log-linear models corresponding to the following nine patterns of allele frequency configurations: 1) shared by all disorders model ([schizophrenia, bipolar I, bipolar II, major depressive disorder]≠control); 2) shared by psychotic disorders model ([schizophrenia, bipolar I]≠[major depressive disorder, bipolar II, control]); 3) shared by mood disorders model ([bipolar I, bipolar II, major depressive disorder]≠[schizophrenia, controls]); 4) shared by depressive disorders model ([bipolar II, major depressive disorder]≠[bipolar I, schizophrenia, control]); 5) schizophrenia-specific (schizophrenia≠[major depressive disorder, bipolar I, bipolar II, control]); 6) bipolar disorder-specific ([bipolar I, bipolar II]≠[major depressive disorder, schizophrenia, control]); 7) bipolar I-specific (bipolar I≠[bipolar II, major depressive disorder, schizophrenia, control]); 8) bipolar II-specific (bipolar II≠[bipolar I, major depressive disorder, schizophrenia, control]); and 9) major depressive disorder-specific (major depressive disorder≠[schizophrenia, bipolar I, bipolar II, control]).
We then identified the best-fit model based on the Bayesian information criterion. Last, in order to con trol the marker-wise type I error at alpha, only the omnibus test p value (4 df) and the best-fit model are reported as primary results. Thus, the result of this analytic procedure is a p value for the single omnibus test and a best-fit model that indicates the phenotype(s) providing the best fit for genotype-phenotype association for a given variant.
As is customary in pooled GWAS analyses, we do not adjust p values for the prior GWAS results of the individual samples.
Results
After quality control, 4,186 unrelated individuals of European ancestry were included in the GWAS analyses in four diagnostic groups: schizophrenia (N=402), bipolar I disorder (N=1,021), bipolar II disorder (N=493), major depressive disorder (N=1,210), and control (N=1,060) (Table 1). A Q-Q plot for the omnibus test is shown in Figure 1. The genomic inflation factor λ was 1.0517.
| Characteristic | Schizophrenia (CATIE [N=402]) | Bipolar I Disorder (STEP-BD [N=1,021]) | Bipolar II Disorder (STEP-BD [N=493]) | Major Depressive Disorder (STAR*D [N=1,210]) | Comparison (NIMH [N=1,060]) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | N | % | |
| Female | 93 | 23.1 | 544 | 53.3 | 295 | 59.8 | 709 | 58.6 | 497 | 46.9 |
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Age (years) | 41.3 | 11.4 | 43.0 | 12.7 | 42.8 | 12.6 | 42.9 | 13.6 | 51.7 | 17.4 |
| Age at onset (years) | 26.8 | 9.2 | 17.6 | 8.9 | 16.7 | 8.7 | 25.5 | 14.7 | N/A | N/A |
TABLE 1. Demographic Characteristics of Individuals of European Ancestry in GWAS Analysesa
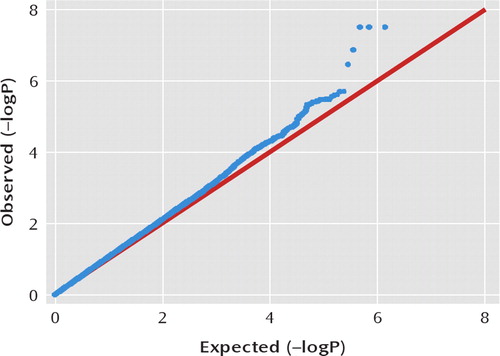
FIGURE 1. Q-Q Plot for the Genomewide Omnibus Test
Genomewide Analysis and Phenotypic Model Selection
SNPs for which the omnibus p value was <5×10-5 are summarized in Table 2, according to the best-fitting model identified by the Bayesian information criterion. A total of 124 SNPs in 25 independent regions exceeded this threshold. For each gene region (positions based on the University of California Santa Cruz genome build hg18), Table 2 lists the number of SNPs in linkage disequilibrium (R2≥0.8) with the top SNP that exceeded the omnibus p value threshold of <5×10-5 (for complete results, see Table 1 in the data supplement accompanying the online version of this article).
| SNP | Chromosome | Top SNP | Alleleb | Gene | Position | Frequencyc | Best-Fit Model | OMNIBUS p | Genotyped or Imputed SNP | Genotyped pd | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bipolar Disorder I | Bipolar Disorder II | Schizophrenia | Major Depressive Disorder | Control | ||||||||||
| 3 | 11 | rs648218 | A/G | ADM | 61 kb down | 0.13 | 0.07 | 0.13 | 0.13 | 0.14 | Bipolar II specific | 3.93E-08 | Genotyped | 3.93E-08 |
| 3 | 22 | rs1001021 | A/G | MYO18B | intron 42 | 0.03 | 0.05 | 0.05 | 0.05 | 0.02 | All disorders | 2.39E-06 | Genotyped | 2.39E-06 |
| 16 | 13 | rs7326068 | A/G | IFT88 | intron 16 | 0.18 | 0.23 | 0.16 | 0.23 | 0.19 | Depressive disorders | 2.92E-06 | Genotyped | 2.92E-06 |
| 1 | 9 | rs3758354 | C/A | ANXA1 | 8 kb up | 0.05 | 0.08 | 0.04 | 0.04 | 0.03 | Bipolar II specific | 3.31E-06 | Genotyped | 3.31E-06 |
| 12 | 14 | rs4982029 | A/G | NPAS3 | intron 1 | 0.04 | 0.04 | 0.04 | 0.03 | 0.02 | All disorders | 3.96E-06 | Imputed | 4.48E-06 |
| 13 | 18 | rs11875674 | C/T | TXNL1 | 153 kb up | 0.22 | 0.27 | 0.21 | 0.19 | 0.24 | Bipolar II specific | 3.97E-06 | Imputed | 1.81E-05 |
| 6 | 1 | rs4271171 | T/C | FAM20B | 8 kb up | 0.53 | 0.58 | 0.49 | 0.54 | 0.50 | Mood disorders | 4.12E-06 | Imputed | 1.16E-05 |
| 1 | 7 | rs12539410 | C/G | CHCHD3 | 403 kb up | 0.01 | 0.03 | 0.01 | 0.02 | 0.01 | Depressive disorders | 4.74E-06 | Imputed | |
| 1 | 7 | rs4726220 | T/C | ACTR3B | 129 kb down | 0.07 | 0.04 | 0.07 | 0.08 | 0.08 | Bipolar II specific | 1.16E-05 | Genotyped | 1.16E-05 |
| 2 | 1 | rs17014011 | A/T | intergenic | 0.13 | 0.15 | 0.16 | 0.15 | 0.11 | All disorders | 1.74E-05 | Imputed | 2.36E-05 | |
| 4 | 2 | rs2372008 | G/A | CRIM1 | 782 kb up | 0.44 | 0.38 | 0.45 | 0.38 | 0.41 | Psychotic disorders | 1.78E-05 | Genotyped | 1.78E-05 |
| 4 | 10 | rs7071307 | A/C | PPP2R2D | 360 kb up | 0.43 | 0.49 | 0.46 | 0.42 | 0.42 | Bipolar II specific | 1.95E-05 | Imputed | 1.98E-05 |
| 1 | 5 | rs194487 | T/C | CTNND2 | intron 13 | 0.32 | 0.34 | 0.30 | 0.35 | 0.28 | Mood disorders | 2.34E-05 | Imputed | |
| 2 | 2 | rs278865 | A/G | NR4A2 | 760 kb up | 0.32 | 0.30 | 0.26 | 0.33 | 0.35 | Schizophrenia specific | 2.46E-05 | Imputed | |
| 7 | 10 | rs10508451 | T/C | OPTN | 137 kb up | 0.35 | 0.30 | 0.40 | 0.38 | 0.35 | Bipolar II specific | 2.87E-05 | Genotyped | 2.87E-05 |
| 2 | 18 | rs12458992 | G/A | CCDC102B | 400 kb up | 0.31 | 0.37 | 0.34 | 0.37 | 0.33 | Depressive disorders | 3.59E-05 | Imputed | |
| 1 | 15 | rs16949856 | T/C | intergenic | 0.26 | 0.22 | 0.26 | 0.21 | 0.26 | Depressive disorders | 3.62E-05 | Imputed | ||
| 1 | 15 | rs2117975 | A/G | EIF2AK4 | 91 kb up | 0.15 | 0.11 | 0.10 | 0.15 | 0.15 | Schizophrenia specific | 3.70E-05 | Imputed | |
| 10 | 20 | rs6079501 | G/T | SEL1L2 | 739 kb down | 0.31 | 0.34 | 0.28 | 0.31 | 0.27 | Mood disorders | 3.73E-05 | Imputed | 3.82E-05 |
| 4 | 12 | rs4140862 | A/C | TMEM16B | intron 5 | 0.30 | 0.33 | 0.33 | 0.36 | 0.36 | Psychotic disorders | 4.03E-05 | Imputed | 4.84E-05 |
| 1 | 7 | rs917815 | G/A | SP8 | 270 kb down | 0.40 | 0.32 | 0.35 | 0.40 | 0.40 | Bipolar II specific | 4.18E-05 | Genotyped | 4.18E-05 |
| 1 | 3 | rs9819616 | G/A | RBMS3 | 294 kb down | 0.35 | 0.34 | 0.39 | 0.31 | 0.36 | Major depressive disorder specifi c | 4.46E-05 | Imputed | |
| 2 | 11 | rs11237798 | G/A | ODZ4 | 541 kb down | 0.08 | 0.08 | 0.11 | 0.09 | 0.12 | Mood disorders | 4.54E-05 | Imputed | |
| 1 | 21 | rs7280842 | T/C | SAMSN1 | 176 kb down | 0.15 | 0.18 | 0.19 | 0.18 | 0.14 | All disorders | 4.56E-05 | Genotyped | 4.56E-05 |
| 2 | 4 | rs7678609 | A/G | HAR39 | 26 kb down | 0.36 | 0.32 | 0.33 | 0.34 | 0.29 | All disorders | 4.58E-05 | Imputed | |
TABLE 2. SNPs Reaching the Omnibus GWAS Test Threshold Among Individuals of European Ancestrya
One region (consisting of seven SNPs) on chromosome 11p15.4 exceeded a genomewide significant threshold for association (Figure 2). The top SNP achieving a p value of 3.93×10-8 was the genotyped SNP rs6484218. Two other SNPs, rs10770107 and rs7119983, also reached a genome-wide significance level of 4.07×10-8 and 4.22×10-8, respectively. These SNPs are located approximately 60 kb from the 3′ end of the nearest gene, adrenomedullin (ADM), and within an expressed sequence tag, EF537581, about which little is known. The best-fitting model for these SNPs indicated effects specific to bipolar II disorder. As shown in Table 2, the minor (A) allele of rs6484218 was less common in the bipolar II disorder group relative to all other groups. Further support for the bipolar II specific model is depicted in Figure 3 of the data supplement, which plots the Bayesian information criterion for all 10 models at rs6484218. As shown in the data supplement, the Bayesian information criterion for the bipolar II disorder-only model is substantially lower than that of all other models.
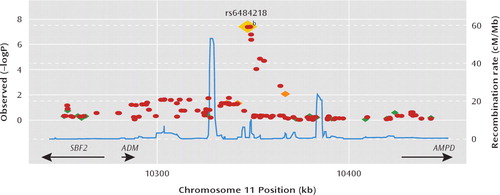
FIGURE 2. Region of Strongest SNP Associationa
aResults (–log10P ) are shown for directly genotyped (diamonds) and imputed (circles) SNPs. The most associated SNP is shown in dark red, and the color of the remaining markers reflects the linkage disequilibrium (R2) with the top SNP in each panel (increasing red hue associated with increasing (R2). The recombination rate (second y axis) is plotted in light blue and is based on the Centre d'Etude du Polymorphisme Humain from Utah HapMap population.
bp=3.93e–08.
The region showing the second strongest evidence of association consisted of three SNPs within intron 42 of the myosin XVIIIB (MYO18B) gene. The top genotyped SNP, rs1001021, showed a p value of 2.39×10-6, accompanied by two imputed SNPs, rs5752249 and rs16986621, with p values of <3.12×10-6. The best-fitting model (the all disorders model) indicated that these SNPs have pleiotropic effects influencing all three disorders. Another region showing the same pleiotropic effects across the three disorders consisted of 12 SNPs within intron 1 of the neuronal PAS domain 3 (NPAS3) gene (top SNP: rs4982029; p=3.96×10-6), a gene previously implicated in psychotic and mood disorders (36–38). Three of the 12 SNPs were directly geno-typed (rs7142052, rs10483422, and rs4982031; all p values <9×10-6 [see Table 1 in the data supplement]).
Also of note, our model selection procedure suggested that genetic variants have effects that differ in their specificity with regard to diagnostic categories. For example, results for five of the 25 regions shown in Table 2 fit best with a model in which they have effects across schizophrenia, bipolar disorder, and major depressive disorder, while four were most consistent with effects on the broad category of mood disorders (bipolar disorder and major depressive disorder) and 10 appeared to fit best with a single disorder.
Discussion
We combined genomewide association methods and a model selection procedure to examine the specificity of genetic influences on three major mental illnesses: schizophrenia, bipolar disorder, and major depressive disorder. This study is the first, to our knowledge, to systematically examine genetic effects across these disorders in a genomewide framework. We observed genomewide significant evidence of association for SNPs near the ADM gene on chromosome 11p. The best-fitting model for our strongest single marker results indicated effects specific to bipolar II disorder. Family studies have supported the hypothesis that bipolar I and II disorder are at least partly genetically distinct (1). Risks of bipolar II disorder tend to be highest among relatives of bipolar II disorder probands as opposed to those with bipolar I disorder or major depressive disorder (39–44). Twin data support the heritability of bipolar II disorder but also suggest shared genetic influences with bipolar I disorder (45). Linkage analyses of bipolar II disorder have found modest evidence of linkage on chromosomes 9p and 18q21 (46). Interestingly, the sixth-ranked region (top SNP: rs11875674; p=3.97×10-6) in our GWAS is located on 18q21, and model selection implicates a bipolar II disorder effect, although this is 7 Mb from the reported linkage peak.
Our results are the first evidence, to our knowledge, implicating specific genetic variants in the risk for bipolar II disorder and provide support for the hypothesis that the genetic etiology of this disorder is distinguishable from other DSM-IV mood disorders. The genomewide signifi-cant signals we observed are located within an expressed sequence tag (EF537581) that has not been well-characterized. This expressed sequence tag does not appear to be brain-expressed (47), making it an implausible candidate for bipolar II disorder. The nearest gene to the associated SNPs is ADM, the gene encoding adrenomedullin. Adrenomedullin is widely expressed in the brain, and mice lacking CNS adrenomedullin exhibit hyperactivity, increased anxiety, and increased sensitivity to the neurotoxic effects of hypobaric hypoxia (48). Elevations of serum adrenomedullin have been reported in bipolar disorder (49), and a recent study identified a functional ADM variant (rs11042725) that was associated with self-reported response to paroxetine among patients with depression (50). We were unable to examine this SNP (or proxies for it) because it is not included in our GWAS or in the HapMap database. However, the SNPs for which we found association do not appear to be in strong linkage disequilibrium with SNPs in adrenomedullin.
Our results also provide preliminary evidence that several SNPs, including markers within several gene regions, have pleiotropic effects that cross traditional DSM-IV boundaries. Although these results did not reach genomewide thresholds for statistical significance, they are consistent with emerging evidence of shared genetic influences on psychotic and mood disorders. In particular, we note a group of SNPs in intron 1 of NPAS3 (top SNP with a p value of 3.96×10-6), for which the best-fitting model indicated effects on schizophrenia, bipolar disorder, and major depressive disorder. The NPAS3 gene encodes a neuronal transcription factor and was identified as a candidate locus for schizophrenia after a balanced translocation (t [9, 14][q34.2;q13]) disrupting the gene was observed to segregate with schizophrenia and learning disability (37). Mice deficient for NPAS3 display behavioral abnormalities, including altered prepulse inhibition, impaired recognition memory, and dysregulation of glutamatergic, dopaminergic, and serotoninergic neurotransmission as well as diminished hippocampal neurogenesis (51–53). Pickard et al. (38) recently reported evidence that haplotypes of NPAS3 are associated with both schizophrenia and bipolar disorder, although the SNPs reported in Table 2 of the present study are not in linkage disequilibrium with those reported by Pickard et al. Nevertheless, our evidence that variation in NPAS3 may have pleiotropic effects on mood and psychotic disorders is intriguing, given the diverse neurobiologic functions of this gene.
Our results have several implications for future genetic studies of major mental illness. First, the analytic approach illustrated provides a basis for identifying genes that underlie the common genetic basis of schizophrenia, bipolar disorder, and major depressive disorder. It is likely that genes influencing these disorders consist of a combination of disorder-specific and shared susceptibility loci. Similar evidence for cross-disorder genetic effects have emerged from GWAS of other medical disorders, most notably autoimmune diseases where specific genetic markers are strongly associated with multiple, clinically distinctive conditions (54, 55). For example, nearly one-half of the risk alleles identified as influencing either type I diabetes or celiac disease have been shown to influence both disorders (55, 56). In addition, GWAS have demonstrated that a coding SNP in the PTPN22 gene is associated with reduced risk of Crohn's disease but increased risk of systemic lupus erythematosis, rheumatoid arthritis, type I diabetes, and Graves disease (54). Second, our results may inform case definition in future genetic studies of schizophrenia, bipolar disorder, and major depressive disorder. Specifically, the power of future analyses may be enhanced by combining affecteds across disorders for loci that appear to have cross-disorder effects. Finally, identifying susceptibility loci that have pleiotropic effects may reveal shared etiologic mechanisms underlying multiple diseases. Biological pathways involving these loci may represent targets for the development of broad-spectrum treatments with efficacy for a wider range of psychopathology than currently available options. We note that the STAR*D sample excluded patients with psychotic depression. While this would not bias our results, it might have implications for their generalizability. To the extent that psychosis itself may be under genetic influence, this exclusion might have made the diagnostic groups more distinct than they would have been had psychosis been present in the major depressive disorder group.
Our results should be considered in light of several limitations. First, given the available sample size, our study had limited power to detect modest genetic effects or rare susceptibility variants using a strict genomewide threshold for significance. Because this is the first report of its kind and the goal was to characterize genotype-phenotype relationships, we present results down to a threshold of p<5×10-5 to inform future analyses. Although our study detected only one region exceeding genomewide significant thresholds, recent pooled analyses of GWAS data have demonstrated that many variants that fall short of this threshold in a single study emerge as confirmed susceptibility loci when additional data sets are analyzed (20, 21). Thus, the top hits in our study provide preliminary evidence that awaits replication in future studies. A large-scale effort to use genomewide data to dissect within-disorder and cross-disorder influences has begun through the Psychiatric GWAS Consortium (57). Our results may inform these analyses, which ultimately may include GWAS data on >80,000 individuals across five disorders: schizophrenia, bipolar disorder, major depressive disorder, autism, and attention deficit hyperactivity disorder (57).
A second limitation is that by design, our model fitting approach did not involve statistical testing of each model versus the null hypothesis. Instead, to control inflation of type I error, we performed only the single omnibus test followed by model selection. A single test of the true underlying model would be expected to have greater power than the omnibus test. However, since the true model is unknown a priori, we would have to test multiple models to cover the range of possible true models. We have shown by simulation that the omnibus test has power comparable to a test of the true model, especially when the latter is corrected for multiple testing (unpublished data available upon request from Purcell et al.).
In summary, in a genomewide association analysis of three major psychiatric disorders, we identified a region of genomewide significant association on chromosome 11p15, near the ADM gene, which appears to be specific to bipolar II disorder. We also found preliminary evidence for both cross-disorder and disorder-specific influences on schizophrenia, bipolar disorder, and major depressive disorder. Consistent with previous reports in independent samples, we observed evidence that variation in NPAS3 has effects that transcend DSM-IV diagnostic categories. The methods illustrated here may inform larger-scale efforts to examine cross-disorder genetic effects.
1. : Family, twin, and adoption studies of bipolar disorder. Am J Med Genet C Semin Med Genet 2003; 123:48–58Crossref, Google Scholar
2. : Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry 2000; 157:1552–1562Link, Google Scholar
3. : The genetics of schizophrenia. PLoS Med 2005; 2:e212Crossref, Medline, Google Scholar
4. : Morbidity risks of schizophrenia and affective disorders among first degree relatives of patients with schizophrenia, mania, depression and surgical conditions. Br J Psychiatry 1980; 137:497–504Crossref, Medline, Google Scholar
5. : Are schizophrenia and affective disorder related? Preliminary data from a family study. Am J Psychiatry 1993; 150:278–285Link, Google Scholar
6. : Increased morbid risk for schizophrenia in families of in-patients with bipolar illness. Schizophr Res 2000; 42:83–90Crossref, Medline, Google Scholar
7. : Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet 2009; 373:234–239Crossref, Medline, Google Scholar
8. : A twin study of genetic relationships between psychotic symptoms. Am J Psychiatry 2002; 159:539–545Link, Google Scholar
9. : The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch Gen Psychiatry 2003; 60:497–502Crossref, Medline, Google Scholar
10. : Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry 2002; 7:405–411Crossref, Medline, Google Scholar
11. : Genes for schizophrenia and bipolar disorder? Implications for psychiatric nosology. Schizophr Bull 2006; 32:9–16Crossref, Medline, Google Scholar
12. : Combined analysis from eleven linkage studies of bipolar disorder provides strong evidence of susceptibility loci on chromosomes 6q and 8q. Am J Hum Genet 2005; 77:582–595Crossref, Medline, Google Scholar
13. : Schizophrenia and affective disorders-cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 fi ndings in a family. Am J Hum Genet 2001; 69:428–433Crossref, Medline, Google Scholar
14. : The schizophrenia phenotype in 22q11 deletion syndrome. Am J Psychiatry 2003; 160:1580–1586Link, Google Scholar
15. : Clinical features of 78 adults with 22q11: deletion syndrome. Am J Med Genet A 2005; 138:307–313Crossref, Medline, Google Scholar
16. : Genotype-phenotype studies in bipolar disorder showing association between the DAOA/G30 locus and persecutory delusions: a first step toward a molecular genetic classifi cation of psychiatric phenotypes. Am J Psychiatry 2005; 162:2101–2108Link, Google Scholar
17. : Variation at the DAOA/G30 locus infl uences susceptibility to major mood episodes but not psychosis in schizophrenia and bipolar disorder. Arch Gen Psychiatry 2006; 63:366–373Crossref, Medline, Google Scholar
18. : No significant association of 14 candidate genes with schizophrenia in a large European ancestry sample: implications for psychiatric genetics. Am J Psychiatry 2008; 165:497–506Link, Google Scholar
19. : Allelic association of G72/G30 with schizophrenia and bipolar disorder: a comprehensive meta-analysis. Schizophr Res 2008; 98:89–97Crossref, Medline, Google Scholar
20. ; Molecular Genetics of Schizophrenia Collaboration: Identifi cation of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet 2008; 40:1053–1055Crossref, Medline, Google Scholar
21. : Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet 2008; 40:1056–1058Crossref, Medline, Google Scholar
22. : Genomewide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Mol Psychiatry 2009; 14:359–375Crossref, Medline, Google Scholar
23. : Gene-wide analyses of genomewide association data sets: evidence for multiple common risk alleles for schizophrenia and bipolar disorder and for overlap in genetic risk. Mol Psychiatry 2009; 14:252–260Crossref, Medline, Google Scholar
24. : Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460:748–752Crossref, Medline, Google Scholar
25. : Rationale, design, and methods of the Systematic Treatment Enhancement Program for Bipolar Disorder (STEP-BD). Biol Psychiatry 2003; 53:1028–1042Crossref, Medline, Google Scholar
26. : Demographic and diagnostic characteristics of the first 1,000 patients enrolled in the Systematic Treatment Enhancement Program for Bipolar Disorder (STEP-BD). Bipolar Disord 2004; 6:460–469Crossref, Medline, Google Scholar
27. : The National Institute of Mental Health Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE) project: schizophrenia trial design and protocol development. Schizophr Bull 2003; 29:15–31Crossref, Medline, Google Scholar
28. : Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. N Engl J Med 2005; 353:1209–1223Crossref, Medline, Google Scholar
29. : Structured Clinical Interview for DSM-IV, Outpatient Version (SCID-OP). New York, Biometrics Research Department, New York State Psychiatric Institute, 1996Google Scholar
30. : Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry 2008; 13:570–584Crossref, Medline, Google Scholar
31. : Sequenced Treatment Alternatives to Relieve Depression (STAR*D): rationale and design. Control Clin Trials 2004; 25:119–142Crossref, Medline, Google Scholar
32. : Whole-genome association study of bipolar disorder. Mol Psychiatry 2008; 13:558–569Crossref, Medline, Google Scholar
33. : Genome-wide association analysis identifi es loci for type 2 diabetes and triglyceride levels. Science 2007; 316:1331–1336Crossref, Medline, Google Scholar
34. : A genomewide association study of citalopram response in major depressive disorder. Biol Psychiatry 2010; 67:133–138Crossref, Medline, Google Scholar
35. : PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81:559–575Crossref, Medline, Google Scholar
36. : Disruption of the neuronal PAS3 gene in a family affected with schizophrenia. J Med Genet 2003; 40:325–332Crossref, Medline, Google Scholar
37. : Disruption of a brain transcription factor, NPAS3, is associated with schizophrenia and learning disability. Am J Med Genet B Neuropsychiatr Genet 2005; 136B:26–32Crossref, Medline, Google Scholar
38. : Interacting haplotypes at the NPAS3 locus alter risk of schizophrenia and bipolar disorder. Mol Psychiatry 2009; 14:874–884Crossref, Medline, Google Scholar
39. : A family study of schizoaffective, bipolar I, bipolar II, unipolar and normal control probands. Arch Gen Psychiatry 1982; 39:1157–1167Crossref, Medline, Google Scholar
40. : A family study of bipolar II disorder. Br J Psychiatry 1984; 145:49–54Crossref, Medline, Google Scholar
41. : Bipolar II: Combine or keep separate? J Affect Disord 1985; 8:17–28Crossref, Medline, Google Scholar
42. : Familial rates of affective disorder: a report from the National Institute of Mental Health Collaborative Study. Arch Gen Psychiatry 1987; 44:461–469Crossref, Medline, Google Scholar
43. : The distinction of bipolar II disorder from bipolar I and recurrent unipolar depression: results of a controlled family study. Acta Psychiatr Scand 1993; 87:279–284Crossref, Medline, Google Scholar
44. : Bipolar II: The most common bipolar phenotype? Am J Psychiatry 1993; 150:901–903Link, Google Scholar
45. : Heritability of bipolar spectrum disorders: Unity or heterogeneity? J Affect Disord 2008; 106:229–240Crossref, Medline, Google Scholar
46. : Genome-wide scan of bipolar II disorder. Bipolar Disord 2007; 9:580–588Crossref, Medline, Google Scholar
47. : Identifi cation of FAM46D as a novel cancer/testis antigen using EST data and serological analysis. Genomics 2009; 94:153–160Crossref, Medline, Google Scholar
48. : Lack of adrenomedullin in the mouse brain results in behavioral changes, anxiety, and lower survival under stress conditions. Proc Natl Acad Sci U S A 2008; 105:12581–12586Crossref, Medline, Google Scholar
49. : Possible role of nitric oxide and adrenomedullin in bipolar affective disorder. Neuropsychobiology 2002; 45:57–61Crossref, Medline, Google Scholar
50. : Association of a functional polymorphism in the adrenomedullin gene (ADM) with response to paroxetine. Pharmacogenomics J 2010; 10:126–133Crossref, Medline, Google Scholar
51. : Abnormal neurodevelopment, neurosignaling and behaviour in NPAS3-deficient mice. Eur J Neurosci 2005; 22:1265–1276Crossref, Medline, Google Scholar
52. : Behavioral and regulatory abnormalities in mice deficient in the NPAS1 and NPAS3 transcription factors. Proc Natl Acad Sci U S A 2004; 101:13648–13653Crossref, Medline, Google Scholar
53. : The neuronal PAS domain protein 3 transcription factor controls FGF-mediated adult hippocampal neurogenesis in mice. Proc Natl Acad Sci U S A 2005; 102:14052–14057Crossref, Medline, Google Scholar
54. : Autoimmune diseases: insights from genome-wide association studies. Hum Mol Genet 2008; 17:R116–R121Crossref, Medline, Google Scholar
55. : Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med 2008; 359:2767–2777Crossref, Medline, Google Scholar
56. : Shared genetic risk factors for type 1 diabetes and celiac disease. N Engl J Med 2008; 359:2837–2838Crossref, Medline, Google Scholar
57.



