Contribution of Intellectual Disability–Related Genes to ADHD Risk and to Locomotor Activity in Drosophila
Abstract
Objective:
Attention deficit hyperactivity disorder (ADHD) is a common, highly heritable neuropsychiatric disorder. ADHD often co-occurs with intellectual disability, and shared overlapping genetics have been suggested. The aim of this study was to identify novel ADHD genes by investigating whether genes carrying rare mutations linked to intellectual disability contribute to ADHD risk through common genetic variants. Validation and characterization of candidates were performed using Drosophila melanogaster.
Methods:
Common genetic variants in a diagnostic gene panel of 396 autosomal intellectual disability genes were tested for association with ADHD risk through gene set and gene-wide analyses, using ADHD meta-analytic data from the Psychiatric Genomics Consortium for discovery (N=19,210) and ADHD data from the Lundbeck Foundation Initiative for Integrative Psychiatric Research for replication (N=37,076). The significant genes were functionally validated and characterized in Drosophila by assessing locomotor activity and sleep upon knockdown of those genes in brain circuits.
Results:
The intellectual disability gene set was significantly associated with ADHD risk in the discovery and replication data sets. The three genes most consistently associated were MEF2C, ST3GAL3, and TRAPPC9. Performing functional characterization of the two evolutionarily conserved genes in Drosophila melanogaster, the authors found that their knockdown in dopaminergic (dMEF2) and circadian neurons (dTRAPPC9) resulted in increased locomotor activity and reduced sleep, concordant with the human phenotype.
Conclusions:
This study reveals that a large set of intellectual disability–related genes contribute to ADHD risk through effects of common alleles. Utilizing this continuity, the authors identified TRAPPC9, MEF2C, and ST3GAL3 as novel ADHD candidate genes. Characterization in Drosophila suggests that TRAPPC9 and MEF2C contribute to ADHD-related behavior through distinct neural substrates.
Attention deficit hyperactivity disorder (ADHD) is a common neurodevelopmental disorder with prevalence estimates of 5.3% in childhood and 2.5–4.9% in adulthood (1). ADHD is clinically characterized by two core symptom domains: inattention and hyperactivity/impulsivity, which can occur individually or in combination (1). Despite the high heritability of ADHD (70–80%) (1), identification of ADHD risk genes has been difficult, mainly because of the disorder’s complex genetic architecture (1). Genetic variants that occur frequently in the population and have generally small individual effects on disease risk are thought to underlie the disorder in most patients, and the first genome-wide significant findings for ADHD have been identified only recently (2).
Intellectual disability refers to a highly heterogeneous group of childhood-onset disorders characterized by below-average intellectual functioning (IQ <70) and significant limitations in adaptive functioning, which covers many everyday social and practical skills (3). Intellectual disability has an estimated prevalence of 2%−3% in the population; severe disability has a population prevalence of 0.3%−0.5% (4). Intellectual disability is often monogenic, but many different genes and types of mutations are implicated (3). ADHD is a common comorbid disorder in children with intellectual disability (5). Studies of children with mild and borderline intellectual disability have identified ADHD in 8%−39% of cases (5). A recent study using Swedish birth registry data showed that nearly all of this comorbidity can be attributed to genetic factors (6). Based on such phenotypic and genetic overlap, it has been hypothesized that intellectual disability and ADHD, and neurodevelopmental disorders more broadly, have an overlapping genetic etiology (6).
In this study, we evaluated the genetic overlap between intellectual disability and ADHD in an attempt to identify novel ADHD candidate genes. We investigated whether genes affected by rare mutations in patients with intellectual disability also contribute to ADHD risk through common genetic variation. For this, we used the latest data freeze from the Psychiatric Genomics Consortium (N=19,210) for discovery and the Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH) sample (N=37,076) for replication. To provide functional support for the newly identified ADHD candidates, we used Drosophila melanogaster, a model that can facilitate characterization of the involved neural substrates. The role of dopaminergic neurotransmission is well established in ADHD (1). In addition, circadian genes and circuits have been implicated, as ADHD often goes together with sleep disturbances, and abnormal circadian rhythms of melatonin secretion have been observed in children and adult patients with ADHD (7). Notably, positive genetic correlations between insomnia and sleep-related traits and ADHD exist (8). Moreover, disrupting the activity of the circadian clock gene Per1 in both mice and zebrafish revealed ADHD-like symptoms (9). In Drosophila, Per1 mutants were found to be deficient for experience-dependent increases in sleep (10). We therefore set out to investigate potential dopaminergic and circadian rhythm components of the identified phenotypes in Drosophila. Dissecting the role of neuronal circuits can help pinpoint the neurotransmitter systems contributing to ADHD as a first step toward individualization of treatment. On down-regulation of gene expression pan-neuronally and in relevant neuronal subsets, we assessed locomotor activity, sleep, and related parameters as behavioral readouts, which we previously established to be relevant for ADHD (11).
Methods
This study used summary statistics of genome-wide association study (GWAS) meta-analyses that had been approved by the local ethics committees and had the required informed consents, as described earlier (2).
Cohorts
The Psychiatric Genomics Consortium (PGC) ADHD GWAS meta-analysis (GWAS-MA) data, which were used at the discovery stage in this study, were available as autosome-wide summary statistics, including single-nucleotide polymorphism (SNP) data with corresponding p values and odds ratios. Data were based on nine studies including 5,621 case subjects and 13,589 control subjects. Samples were of Caucasian or Han Chinese origin and included patients who met DSM-IV criteria for ADHD (see Table S1 in the online supplement). Details on the procedures for DNA isolation, whole-genome genotyping, and imputation have been described previously (12). Briefly, genome-wide data were obtained from different genotyping arrays (see Table S1) and were imputed using 1000 Genomes Project Phase 3 as a reference panel (National Center for Biotechnology Information [NCBI] build 37 [hg19] coordinates) for autosomal SNPs. Meta-analytic data were processed through a stringent quality control pipeline applied at the PGC (12).
The gene set association was replicated in an independent cohort from the iPSYCH–Statens Serum Institut–Broad Institute (iPSYCH-SSI-Broad) ADHD working group (N=37,076) (2).
A meta-analysis of the two data sets described above (20,183 case subjects and 35,191 control subjects) was published recently as part of the ADHD Working Group of the PGC and the ADHD iPSYCH-SSI-Broad collaboration (2). We used this meta-analytic data set to perform a gene-based lookup of three genes of interest, using the Multimarker Analysis of Genomic Annotation (MAGMA) software program, as described below. Detailed quality control and imputation parameters were described in the original publication (2). In short, summary data included only markers that had a quality score (INFO score) >0.8, had a minor allele frequency (MAF) >0.01, and were supported by an effective sample size greater than 70% (8,047,420 markers) (2).
GWAS of ADHD Symptom Scores in the Nijmegen Biomedical Study
From the Nijmegen Biomedical Study, a population-based study in adults (13), data on hyperactivity/impulsivity and inattention symptoms from the self-report DSM-IV-based ADHD Rating Scale (14) and whole-genome genotyping were available for >2,978 individuals (see Table S4 in the online supplement). Detailed information on the sample, procedures of DNA isolation, whole-genome genotyping, and imputation is provided in the Supplementary Methods section in the online supplement. Genome-wide association analysis was performed using a linear regression under an additive model in PLINK, version 1.9 (15, 16), using Ricopili (https://sites.google.com/a/broadinstitute.org/ricopili/). Age, gender, and 10 principal components were included as covariates. For subsequent gene-based analyses, SNPs with an INFO score ≥0.8 and MAF ≥0.01 were included.
Intellectual Disability Gene Selection
For the selection of the intellectual disability gene set, we used the publicly available Intellectual Disability Gene Panel of the Radboud University Medical Center (Radboudumc) Department of Human Genetics’ Genome Diagnostics division (downloaded from https://issuu.com/radboudumc/docs/ngs-intellectual_disability_panel_1?e=28355229/50899368 on March 27, 2014). This gene panel listed 490 intellectual disability–related genes (see Table S2 in the online supplement), based on findings of de novo mutations in patients with intellectual disability visiting the Radboudumc, collaborating institutes, the literature, and public databases. This list forms the basis for diagnostic testing using exome sequencing at Radboudumc.
Gene-Based and Gene Set Analysis
Genome-wide summary statistics of ADHD (PGC and iPSYCH ADHD GWAS-MA) were used as input for gene-based analyses. We used two software packages to test whether the intellectual disability gene set was associated with ADHD risk. First, the hybrid set-based test (HYST) of the Knowledge-Based Mining System for Genome-Wide Genetic studies (KGG), version 3.5 (17), was used for association testing (see the Supplementary Methods section in the online supplement). Second, MAGMA, version 1.02 (18), was used (see the Supplementary Methods section). The analyses were carried out in two steps. In step 1, the combined effect of the SNPs in the vicinity of all intellectual disability genes was analyzed. Post hoc, in step 2, the potential effects of the individual genes were investigated by reviewing their gene-based test statistics. Genes were considered gene-wide significant if they reached the Bonferroni correction threshold adjusted for the number of genes tested (p<0.000128).
Functional Characterization of MEF2C and TRAPPC9 in Drosophila melanogaster
Drosophila strains and breeding.
Drosophila orthologs were retrieved from the NCBI protein DELTA-BLAST and ENSEMBL gene tree (19, 20). The Drosophila orthologs of MEF2C (termed Mef2) and TRAPPC9 (termed brun) were targeted by RNA interference (RNAi)–mediated knockdown using the UAS-GAL4 system. Tissue-cell-type-specific knockdown was achieved using tissue-cell-type-specific promoters driving GAL4 expression. Several neuronal populations were targeted: nSyb-GAL4 (yw* UAS-Dcr-2 hs(X);; nSyb-GAL4 [for Drosophila genotype nomenclature, see https://wiki.flybase.org/wiki/FlyBase:Nomenclature]) (21) targeting all neurons (pan-neuronal driver), tim-GAL4 (tim-GAL4, UAS-Dcr-2/CyO) (22) targeting timeless-expressing cells including circadian neurons, and ple-GAL4 (w*; UAS-Dcr-2; ple-GAL4), obtained from the Bloomington Drosophila Stock Center (stock number 8848), targeting tyrosine hydroxylase–expressing (dopaminergic) neurons, visualized in Figure S1 in the online supplement. A copy of UAS-Dcr-2 was incorporated to improve knockdown efficiency (21). The driver stocks were crossed with UAS-RNAi lines obtained from the Vienna Drosophila Resource Center: v12482 (w1118; UAS-dTRAPPC9RNAi), v15549 (w1118;; UAS-dMEF2RNAi-1), v15550 (w1118;; UAS-dMEF2RNAi-2), and v60000 (w1118). Progeny of the latter crosses served as genetic background controls. Non-induced UAS-RNAi lines were generated by crossing UAS-RNAi stocks with the isogenic line iso31 (Bloomington stock number 5905: w1118), replacing the driver in the cross. The driver lines expression pattern was validated by driving GFP expression and UAS-RNAi lines by quantitative polymerase chain reaction (see the Supplementary Methods section and Figure S1 in the online supplement). All flies were maintained on standard corn meal feed at 28°C with 60% relative humidity in a 12-hour light-dark cycle.
Locomotor activity monitoring and calculation of activity and sleep parameters.
Locomotor activity of individual 3- to 5-day-old male flies was recorded with the Drosophila Activity Monitor system (TriKinetics, Waltham, Mass.). The flies were collected with the aid of CO2 and allowed to recover for 24 hours. The activity count was recorded for 4 days at 28°C and 60% relative humidity in a 12-hour light-dark cycle, followed by 2 days in constant darkness. The activity data were collected every 30 seconds and analyzed in 1-minute bins. Activity and sleep were analyzed with the pySolo software program (23), with sleep defined as ≥5 minutes of inactivity. Average daily activity and sleep were then plotted in 10- and 30-minute bins, respectively. pySolo was modified to analyze total activity and sleep between 180 and 540 minutes zeitgeber time (ZT) for the relative day and between 900 and 1,260 minutes ZT for the relative night to capture periods of stable activity and sleep, as described previously (11). Activity while awake, sleep bout count, sleep bout duration, and sleep latency were extracted using the Sleep and Circadian Analysis MATLAB Program (SCAMP) (24). Data for individual flies from at least two independent experiments were pooled, and t tests were performed, with Welch’s correction when variances were unequal. Results were considered significant if they reached the Bonferroni correction threshold adjusted for the number of drivers tested (p<0.0167). To compare the relative day and night activity, the delta (Δ) activity and sleep between knockdown and genetic background control were calculated (Δday=knockdownday−controlday and Δnight=knockdownnight−controlnight). All statistical analyses were performed with Prism, version 5.03 (GraphPad, San Diego).
Results
Association of Intellectual Disability Gene Set With ADHD Risk
To select candidate genes for the intellectual disability gene set, we used the publicly available Intellectual Disability Gene Panel. Genes were included on the basis of findings of de novo mutations in patients with intellectual disability visiting the Radboudumc and collaborating institutes or reported in the literature and in public databases (N=490) (see Table S2 in the online supplement). The set of intellectual disability genes was tested for association with ADHD by two different software algorithms in a discovery-replication design. For discovery, we used the PGC ADHD GWAS-MA data (N=19,210) and the KGG software program; the HYST test revealed that the intellectual disability gene set as a whole was significantly associated with ADHD risk (pKGG=0.0001; genes, N=387). To assess the robustness of our findings, we tested the association of the intellectual disability gene set with ADHD in the PGC data using MAGMA. The results also showed a significant association of the intellectual disability gene set with ADHD risk in the self-contained test (p=0.0412; genes, N=392), but not in the competitive test (p=0.9522). As an independent replication, we tested the gene set association in the iPSYCH cohort (N=37,076) using MAGMA; the results robustly replicated the significance in the self-contained test (p=1.2429×10−13; genes, N=393). The competitive test was negative again (p=0.5306).
To identify the major contributors to the observed association (i.e., the individual genes most significantly associated) for further validation, we performed individual gene-wide testing within the gene set using the PGC data. The most consistent findings across algorithms were for the Myocyte Enhancer Factor 2C gene (MEF2C; pKGG=1.3×10−5 and pMAGMA=1.497×10−4) (Figure 1A), the Trafficking Protein Particle Complex 9 gene (TRAPPC9; pKGG=7.81×10−7 and pMAGMA=0.0035) (Figure 1B), and the ST3 Beta-Galactoside Alpha–2,3-Sialyltransferase 3 gene (ST3GAL3; pKGG=6.18×10−5 and pMAGMA=6.808×10−4) (Figure 1C). Gene-based p values for all genes in both KGG and MAGMA analyses are listed in Table S3 in the online supplement. A lookup in the recently published combined PGC and iPSYCH GWAS-MA (2) revealed genome-wide significant results for gene-wide analysis of ST3GAL3 and MEF2C, and nominal significance for TRAPPC9 (p values of 4.6406×10−13, 2.671×10−10, and 0.0184, respectively).
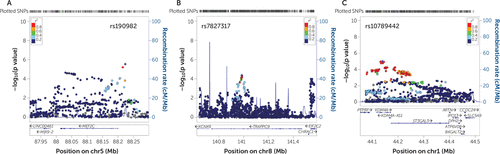
FIGURE 1. Regional association plots showing association signals for ADHD in the Psychiatric Genomics Consortium GWAS meta-analysis (N=19,210) for the three most consistently associated genes, including flanking regions of 100 kba
a Panel A shows the MEF2C locus with the top SNP (rs190982) indicated by the purple dot. Panel B shows the TRAPPC9 locus with the top SNP (rs7827317) indicated by the purple dot. Panel C shows the ST3GAL3 locus with the top SNP (rs10789442) indicated by the purple dot. Results are shown as −log10(p value) for genotyped and imputed SNPs. The color of each marker reflects its linkage disequilibrium (r2) with the strongest associated SNP (in purple). The recombination rate is plotted in blue. chr=chromosome; cM=centimorgan; Mb=megabase.
To distinguish between contributions of the two separate symptom domains of ADHD—hyperactivity/impulsivity and inattention—we investigated gene-based associations in a population-based cohort of >2,978 adults (13). MEF2C and TRAPPC9 showed gene-based association with hyperactive/impulsive ADHD symptoms, but not inattentive ADHD symptoms. However, these results did not survive correction for multiple testing (see Table S5 in the online supplement).
Functional Validation and Characterization of MEF2C and TRAPPC9 in Drosophila
Next, we investigated the validity of the newly identified ADHD candidate genes by mapping their effects on ADHD-related phenotypes in Drosophila melanogaster. As mutations in these genes are already proven to cause monogenic forms of intellectual disability but not ADHD, we focused our efforts on the ADHD-related phenotypes. We did so by investigating neuronal subsets, in addition to pan-neuronal knockdown of the genes. This allowed us to characterize the different circuits through which individual ADHD risk genes may act. Second, it may reveal phenotypes that might otherwise be masked by opposing actions of different neurons in the same circuit (25). We previously established Drosophila as a model for ADHD (11) by showing that pan-neuronal knockdown of ADHD genes preferentially caused (dopamine-related) increased locomotor activity and sleep loss at night. Two of the three ADHD candidate genes were found conserved in Drosophila: the MEF2 gene-family homolog Mef2 (henceforth referred to as dMEF2) and the TRAPPC9 homolog brun (henceforth referred to as dTRAPPC9). While there are four MEF2 genes (A–D) in humans, only a single gene representing the MEF2 family exists in Drosophila, dMEF2. The gene may reflect aspects of the whole human MEF2 family, but nervous system expression is most prominent for MEF2C (see Figure S2 in the online supplement), and studying neural phenotypes of dMEF2 may therefore best model MEF2C. ST3GAL3 is found in vertebrates, and no known ortholog has been identified in Drosophila. We investigated locomotor activity and sleep after knocking down dMEF2 and dTRAPPC9 expression in all neurons, or more specifically in dopaminergic or circadian neurons. Single-cell transcriptome data revealed that dMEF2 and dTRAPPC9 expression can be detected in those specific neuronal populations (see Figure S3 in the online supplement) (26). Cell-type-specific knockdown was achieved by driving the expression of RNAi in the neuronal populations of interest (pan-neuronal, dopaminergic, and circadian) using the binary UAS/Gal4 system. Flies were monitored in a 12-hour light-dark scheme, mimicking a day-and-night period. We also investigated behavior in 24-hour constant darkness conditions, given our earlier model showing that the dopamine-related increased locomotor activity is present in the absence of light (11).
dMEF2 Knockdown Gives Rise to Elevated Nighttime Activity and Sleep Defects
Pan-neuronal knockdown of dMEF2 expression caused no changes in activity and sleep during the stable period of the relative day compared with the genetic background control (Figure 2A), but significantly increased night activity (p=0.0059) and reduced sleep (p=0.014) (Figure 2A; see also Table S6 in the online supplement). In constant darkness, the knockdown also showed significantly increased activity (p=0.0088) and less sleep (p=0.00037) in the relative night period (Figure 2B; see also Table S7 in the online supplement). This increased activity was the result of increased activity counts per waking minute (Figure 2A, B). Further analysis of sleep parameters revealed a tendency of reduced sleep bout duration and an increased sleep latency in the relative night period (see Figure S5A,B in the online supplement). Knockdown of dMEF2 in dopaminergic neurons showed increased night activity (p=1.8×10−15) and reduced sleep (p=5.1×10−15) (Figure 2C; see also Table S6 in the online supplement). Activity and sleep during the relative day period were not different from the genetic background control (Figure 2C). In constant darkness, increased activity and reduced sleep were observed in both relative day (p=4.5×10−7 and p=2.6×10−17, respectively) and night (p=1.6×10−17 and p=9.5×10−26, respectively) (Figure 2D; see also Table S7 in the online supplement). Under these conditions, the increased activity was exclusively driven by a sleep defect; activity while awake was even lower than in controls (Figure 2C,D). The sleep defect was accompanied by an increased sleep bout count and a reduced sleep bout duration (see Figure S5C,D in the online supplement). Knockdown using tim-GAL4 did not yield viable flies, precluding further analysis. The increased activity and sleep loss were the result of induced knockdown of the gene of interest, as non-induced UAS-RNAi lines showed no increased activity or sleep defect (see Figure S7 in the online supplement).
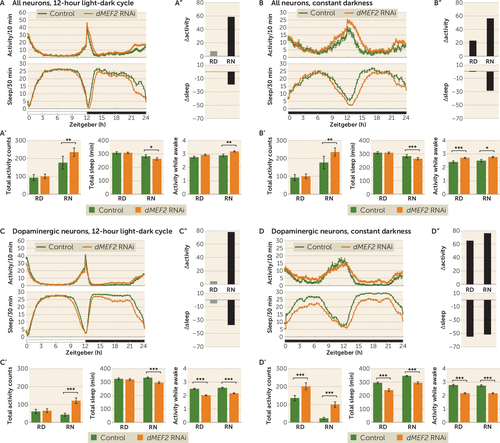
FIGURE 2. Higher activity and reduced sleep in the relative night resulting from knockdown of dMEF2 in all neurons, or in dopaminergic neurons specifically, in Drosophilaa
a Panels A and B present activity and sleep plots of pan-neuronal dMEF2 knockdown in the 12-hour light-dark cycle and in constant darkness, respectively. Panels A′ and B′ present the quantification of total activity, sleep, and activity while awake during stable periods (relative day [RD]: zeitgeber 3–9 hours; relative night [RN]: zeitgeber 15–21 hours), excluding the activity peaks (zeitgeber 0–3 hours, 9–15 hours, and 21–24 hours). Pan-neuronal knockdown of dMEF2 showed increased activity, activity while awake, and sleep loss during the RN period during both the 12-hour light-dark cycle and constant darkness. Panels A′′ and B′′ present Δactivity and Δsleep: the findings for the 12-hour light-dark cycle and for constant darkness both reveal that the difference between groups is greater in the absence of light. Panels C and D present activity and sleep plots of dopaminergic neuron dMEF2 knockdown in the 12-hour light-dark cycle and in constant darkness, respectively. Panels C′ and D′ present the quantification of total activity, sleep, and activity while awake during stable periods (RD: zeitgeber 3–9 hours; RN: zeitgeber 15–21 hours), excluding the activity peaks (zeitgeber 0–3 hours, 9–15 hours, and 21–24 hours). Dopaminergic neuronal knockdown of dMEF2 showed increased activity and sleep loss in the RN period during the 12-hour light-dark cycle and in both RD and RN during constant darkness. The knockdown showed lower activity while awake than the control in both RD and RN in the 12-hour light-dark cycle and also in constant darkness. Panels C′′ and D′′ present Δactivity and Δsleep: the findings for the 12-hour light-dark cycle and for constant darkness reveal that the difference is greater when light is absent. For the figure, data from two dMEF2 lines with identical UAS-RNAi constructs were combined since the results from the individual lines are highly consistent; the individual data are presented in Figure S4, and further activity and sleep parameters in Figure S5, in the online supplement. Error bars represent standard error of the mean. N=3 biological replicates, minimum 20 flies/replicate. *p<0.0167 (Bonferroni correction threshold); **p<0.01; ***p<0.001.
dTRAPPC9 Knockdown Influences Activity and Sleep Only in Neuronal Subtypes
Pan-neuronal knockdown of dTRAPPC9 did not result in observable alterations in activity or sleep either in the 12-hour light-dark cycle or in constant darkness (Figures 3A,B; see also Figure S5A,B in the online supplement). Specific knockdown of dTRAPPC9 in dopaminergic neurons caused significantly reduced activity and increased day sleep during the relative day (p=0.0022 and p=0.013, respectively), but not in the night (Figure 3C; see also Table S6 in the online supplement). In constant darkness, relative night activity was increased and sleep was reduced (p=0.012 and p=0.015, respectively) (Figure 3D; see also Table S7 in the online supplement). In contrast, knockdown of dTRAPPC9 timeless-expressing neurons resulted in increased night activity and reduced night sleep (p=4.2×10−5 and p=0.00022, respectively) (Figure 3E; see also Table S6 in the online supplement). In constant darkness, increased activity and reduced sleep were also present in the relative night (p=0.00017 and p=0.010, respectively) (Figure 3F; see also Table S7 in the online supplement). This increased activity was the result of higher activity counts per waking minute (Figure 3E,F). Further analysis of sleep parameters revealed an increased sleep bout count and an increased sleep latency in the relative night period (see Figure S6E,F in the online supplement). The activity and sleep loss were the result of knockdown of the gene of interest, as non-induced UAS-RNAi did not show increased activity or sleep defects (see Figure S7 in the online supplement).
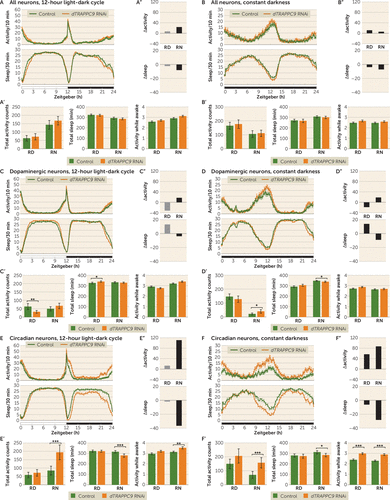
FIGURE 3. Higher activity and reduced sleep resulting from knockdown of dTRAPPC9 when induced in circadian rhythm neurons but not in all or dopaminergic neuronsa
a Panels A and B present activity and sleep plots of pan-neuronal dTRAPPC9 knockdown in the 12-hour light-dark cycle and in constant darkness, respectively. Panels A′ and B′ present the quantification of total activity, sleep, and activity while awake during stable periods (relative day [RD]: zeitgeber 3–9 hours; relative night [RN]: zeitgeber 15–21 hours), excluding the activity peaks (zeitgeber 0–3 hours, 9–15 hours, and 21–24 hours). Pan-neuronal knockdown of dTRAPPC9 showed similar activity, sleep, and activity while awake. Panels A′′ and B′′ present Δactivity and Δsleep. Panels C and D present activity and sleep plots of dopaminergic neuron dTRAPPC9 knockdown in the 12-hour light-dark cycle and in constant darkness, respectively. Panels C′ and D′ present the quantification of total activity, sleep, and activity while awake during stable periods (RD: zeitgeber 3–9 hours; RN: zeitgeber 15–21 hours), excluding the activity peaks (zeitgeber 0–3 hours, 9–15 hours, and 21–24 hours). Dopaminergic neuronal knockdown of dTRAPPC9 showed lower activity and increased sleep in the RD period during the 12-hour light-dark cycle. During constant darkness, the knockdown showed higher activity and sleep loss in the RN period. Panels C′′ and D′′ present Δactivity and Δsleep. Panels E and F present activity and sleep plots of circadian rhythm neuron dTRAPPC9 knockdown in the 12-hour light-dark cycle and in constant darkness, respectively. Panels E′ and F′ present the quantification of total activity, sleep, and activity while awake during stable periods (RD: zeitgeber 3–9 hours; RN: zeitgeber 15–21 hours), excluding the activity peaks (zeitgeber 0–3 hours, 9–15 hours, and 21–24 hours). Circadian neuronal knockdown of dTRAPPC9 showed increased activity and sleep loss in the RN period during both the 12-hour light-dark cycle and constant darkness. The knockdown showed increased activity while awake in the RN period during the 12-hour light-dark cycle and in both the RD and RN periods in constant darkness. Panels E′′ and F′′ present Δactivity and Δsleep: the findings for 12-hour light-dark cycle and for constant darkness reveal that the difference is greater when light is absent. Further activity and sleep parameters are shown in Figure S6 in the online supplement. Error bars represent standard error of the mean. N=3 biological replicates, minimum 20 flies/replicate. *p<0.0167 (Bonferroni correction threshold); **p<0.01; ***p<0.001.
Discussion
In this study, we used a robust discovery-replication design in the currently largest available independent data sets to show that genes affected by rare genetic variation in patients with intellectual disability also contribute to ADHD risk through common genetic variation. In the discovery phase, we also used different algorithms to test gene set association to further test the robustness of findings. In the KGG HYST test and MAGMA, we found significance in both self-contained tests but not the competitive test. A nonsignificant competitive p value in the competitive test should be interpreted as an inability to disentangle the part of the polygenicity attributable to the genes in the gene set from the polygenicity “remaining” (i.e., not captured by the set) on the rest of the genome. In combination with a significance in the self-contained test, it should not be interpreted as no effect of the selected gene set on the outcome. Our replication in the larger independent data set makes this point convincingly. Even more convincing is the fact that two of the three novel ADHD candidate genes that we identified, MEF2C and ST3GAL3, are among the genome-wide significant findings in the recently published ADHD GWAS-MA (2).
Interestingly, our study design produced reproducible findings in much smaller sample sizes than those needed to reach genome-wide significance, which makes such overlap studies an attractive source of genes that have not previously been implicated by GWAS in ADHD. While we based our selection of intellectual disability genes on a diagnostic gene panel, many more intellectual disability genes are currently being discovered through the rapid advances in next-generation sequencing technology; those surely leave additional ADHD genes to be identified.
Our interdisciplinary approach, combining highly powered statistical analyses in humans with functional analyses in an unconventional, validated Drosophila model for ADHD-related behavior (11, 27) allowed for a direct validation and further characterization of neural substrates involved. None of our top three genes had been investigated in the context of ADHD before. MEF2C encodes a member of the MADS box transcription factor, which binds to the conserved MADS box sequence motif (28). MEF2C is important for normal neuronal function by regulating neuronal proliferation, differentiation, survival, and synapse development (29, 30). It also plays a role in hippocampal-dependent learning and memory, possibly by controlling the number of excitatory synapses (31). While both haploinsufficiency and gene duplications of MEF2C give rise to intellectual disability in humans, most severe cases of intellectual disability are linked to large deletions removing part or all of MEF2C and de novo point mutations in the gene (32); individuals with duplications of MEF2C usually display a milder phenotype, with only mild cognitive impairment (33). This is why we chose to model reduced gene expression in Drosophila in this study. Common variants (SNPs) in the MEF2C locus have previously been found to be associated with various cognitive, neuropsychiatric, and neurodegenerative phenotypes, such as intelligence (34), schizophrenia (35), and Alzheimer’s disease (36), indicating pleiotropic effects of this gene on a range of phenotypes. The findings of our study add ADHD to this list and suggest that this is linked to the role of MEF2C in neurotransmission, contributing to it through dopaminergic neurons. However, knowing that in Drosophila dMEF2 expression is important in maintaining normal circadian rhythm (37, 38), we cannot yet rule out an additional role of dMEF2 in circadian neurons in the ADHD-related behaviors, as our dMEF2 knockdown did not yield flies. This could be a result of dMEF2 knockdown in circadian neurons causing cell death. Previous studies on circadian neuron ablation have indeed shown that this leads to embryonic lethality (39). However, since tim-Gal4 has been shown to be expressed outside the nervous system in the fat body and in oenocytes (40, 41), we cannot exclude the possibility that dMEF2 causes this lethality in noncircadian neurons.
In humans, four MEF2 genes (A–D) exist, whereas in Drosophila only a single gene represents the MEF2 family (dMEF2). In this study, we investigated the neuronal function of dMEF2 as a model of human MEF2C function and associated phenotypes, since we found that genetic variation in MEF2C is primarily associated with intellectual disability, whereas genetic variation in MEF2A is mainly linked to coronary artery disease and myocardial infarction, and no clinically relevant entries were found for MEF2D (42). TRAPPC9 has been implicated in NF-kB signaling and is possibly involved in intracellular trafficking. TRAPPC9 is highly expressed in postmitotic neurons of the cerebral cortex, and MRI analysis of affected patients showed defects in axonal connectivity (43). The Drosophila TRAPPC9 has been studied for its involvement in meiotic division in Drosophila male gametes (44), but a neuronal function has not been described so far. TRAPPC9-associated intellectual disability is linked to loss of function of the gene (45). Hyperactive behavior has so far been reported in one patient with a TRAPPC9 mutation (46). Our findings indicate that TRAPPC9 can play a role in ADHD and suggest that the gene acts primarily by affecting neurons involved in circadian regulation. Interestingly, while the dTRAPPC9 dopaminergic neuron knockdown showed similar night activity and sleep profile to that of the control, somewhat lower activity and increased sleep were observed during the day, suggesting that dTRAPPC9 has several cell-type-specific roles in the brain.
While both dMEF2 and dTRAPPC9 pan-neuronal knockdown showed weak or no activity nor sleep phenotype compared with the control, the knockdown in specific neuronal populations did result in pronounced alterations in activity and sleep profiles. Several explanations are possible for the observation of a more pronounced phenotype with specific neuronal subtype knockdown than with a pan-neuronal knockdown; the two most likely ones are 1) that a cell-type-specific driver is stronger in the specific neurons compared with a pan-neuronal driver, and 2) that a targeted gene functions in different circuits controlling the behavior of interest (e.g., excitatory versus inhibitory) that are differentially affected if a cell-type-specific knockdown is performed. Notably, in part of the Drosophila brain, Sitaraman et al. (25) identified distinct neuronal subtypes that oppositely regulate sleep. Considering that the whole nervous system is a mixture of various neuronal populations, each with a specific function in specified phases of development, the different activity and sleep profiles of dMEF2 and dTRAPPC9 knockdown in distinct neuronal populations indicates the need to investigate gene function in different brain circuits and identifies a particular strength of our study. A third possibility to explain the phenotypic differences between lines are differences in the developmental timing of knockdown between the genotypes, as determined by the expression of the utilized drivers. For the latter, nSyb is known to be expressed earlier in development than ple and tim (47); while this does not readily support timing of driver-determined expression as a cause of the different phenotypes, the possibility cannot be excluded. Importantly, our findings also show that different behavioral characteristics can contribute to ADHD-like activity phenotypes downstream of different gene defects; the dMEF2 knockdown showed increased activity and sleep loss as a result of sleep defects, while in dTRAPPC9 knockdown flies, the altered activity and sleep were caused by hyperactivity. Future studies will provide a more in-depth characterization of the neuronal morphology upon dMEF2 and dTRAPPC9 knockdown. Such studies are needed in order to better understand the impact that RNAi has on the morphology and functionality of the relevant neurons. There are several possible explanations for why the observed phenotype arises upon dMEF2 and dTRAPPC9 knockdown: loss of neurons, affected function of these neurons as a result of illness, or disruption of a more specific neuronal process. A similar phenotype was even reported to be caused by hyperactivation of dopaminergic neurotransmission (48) and not by loss of dopaminergic neurons (11). However, the ablation of circadian neurons does cause loss of circadian rhythm (39, 49), which we did not observe upon dTRAPPC9 knockdown. Although neuronal loss might still be a possibility, it is most likely that the phenotype is a result of cellular disturbances at the functional level.
The third ADHD candidate we identified, ST3GAL3, is not conserved in Drosophila, so we were not able to study its contribution to ADHD-relevant behavior. The gene encodes a membrane protein (ST3GalIII) that adds sialic acid to the terminal site of glycolipids or glycoproteins. The gene is expressed in a variety of tissues, including neurons (50). In mice, St3gal2 and St3gal3 are responsible for nearly all the terminal sialylation of brain gangliosides and play an important role in cognition (50). A role in brain development is also likely in humans, as the human brain is particularly enriched in sialic acid–containing glycolipids (i.e., gangliosides) (51). Gangliosides are known to modulate calcium homeostasis and signal transduction in neurons (52). Common genetic variants in ST3GAL3 have also been associated with educational attainment (53). Interestingly, in a recent study of DNA methylation, sites annotated to ST3GAL3 were found to be associated with ADHD symptom trajectories in the population (54). The use of alternative animal models, such as mouse or zebrafish, is warranted to characterize the neuronal circuits underlying ST3GAL3’s effects on ADHD-related behavior.
In this study, we modeled one of the two behavioral symptom domains of ADHD, namely, hyperactivity. This was consistent with our findings—although only nominally significant, likely because of limited sample size—that MEF2C and TRAPPC9 were more strongly associated with hyperactivity/impulsivity than with inattention. However, being able to assess gene effects related to inattention would likely help to elucidate additional neural substrates and circuits involved in ADHD. Currently, multiple paradigms are available to assess attention in Drosophila, as summarized by de Bivort and van Swinderen (55).
In summary, the genetic overlap we observed between intellectual disability and ADHD may suggest biological pleiotropy, in which genetic variation severity in an overlapping set of genes is linked to the severity of neurodevelopmental phenotypes. Functional characterization of neural substrates involved revealed that the novel ADHD candidate genes may affect disease etiology through different biological pathways.
1 : Attention-deficit/hyperactivity disorder. Nat Rev Dis Primers 2015; 1:15020Crossref, Medline, Google Scholar
2 : Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet 2019; 51:63–75Crossref, Medline, Google Scholar
3 : Genetic and epigenetic networks in intellectual disabilities. Annu Rev Genet 2011; 45:81–104Crossref, Medline, Google Scholar
4 : Mental health surveillance among children: United States, 2005–2011. MMWR Suppl 2013; 62:1–35Medline, Google Scholar
5 : Mental disorders in five-year-old children with or without developmental delay: focus on ADHD. J Clinical Child Adolesc Psychol 2010; 39:492–505Crossref, Medline, Google Scholar
6 : The familial co-aggregation of attention-deficit/hyperactivity disorder and intellectual disability: a register-based family study. J Am Acad Child Adolesc Psychiatry 2017; 56:167–174.e1Crossref, Medline, Google Scholar
7 : Adult attention-deficit hyperactivity disorder is associated with alterations in circadian rhythms at the behavioural, endocrine, and molecular levels. Mol Psychiatry 2012; 17:988–995Crossref, Medline, Google Scholar
8 : Biological and clinical insights from genetics of insomnia symptoms. Nat Genet 2019; 51:387–393Crossref, Medline, Google Scholar
9 : Circadian modulation of dopamine levels and dopaminergic neuron development contributes to attention deficiency and hyperactive behavior. J Neurosci 2015; 35:2572–2587Crossref, Medline, Google Scholar
10 : Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science 2009; 324:105–108Crossref, Medline, Google Scholar
11 : ADHD-associated dopamine transporter, latrophilin, and neurofibromin share a dopamine-related locomotor signature in Drosophila. Mol Psychiatry 2016; 21:565–573Crossref, Medline, Google Scholar
12 : Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry 2010; 49:884–897Crossref, Medline, Google Scholar
13 : Cohort profile: the Nijmegen Biomedical Study (NBS). Int J Epidemiol 2017; 46:1099–1100jMedline, Google Scholar
14 : Reliability, validity, and utility of instruments for self-report and informant report concerning symptoms of ADHD in adult patients. J Atten Disord 2008; 11:445–458Crossref, Medline, Google Scholar
15 : Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015; 4:7Crossref, Medline, Google Scholar
16 : PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81:559–575Crossref, Medline, Google Scholar
17 : A knowledge-based weighting framework to boost the power of genome-wide association studies. PLoS One 2010; 5:
18 : MAGMA: generalized gene set analysis of GWAS data. PLOS Comput Biol 2015; 11:
19 : Domain enhanced lookup time accelerated BLAST. Biol Direct 2012; 7:12Crossref, Medline, Google Scholar
20 : Ensembl 2016. Nucleic Acids Res 2016; 44(D1):D710–D716Crossref, Medline, Google Scholar
21 : A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 2007; 448:151–156Crossref, Medline, Google Scholar
22 : A new in vivo model of pantothenate kinase-associated neurodegeneration reveals a surprising role for transcriptional regulation in pathogenesis. Front Cell Neurosci 2013; 7:146Medline, Google Scholar
23 : pySolo: a complete suite for sleep analysis in Drosophila. Bioinformatics 2009; 25:1466–1467Crossref, Medline, Google Scholar
24 : High-resolution positional tracking for long-term analysis of Drosophila sleep and locomotion using the “Tracker” program. PLoS One 2012; 7:
25 : Control of sleep by dopaminergic inputs to the Drosophila mushroom body. Front Neural Circuits 2015; 9:73Medline, Google Scholar
26 : A single-cell transcriptome atlas of the aging drosophila brain. Cell 2018; 174:982–998.e20Crossref, Medline, Google Scholar
27 : Converging evidence does not support GIT1 as an ADHD risk gene. Am J Med Genet B Neuropsychiatr Genet 2015; 168:492–507Crossref, Medline, Google Scholar
28 : Functional regulatory regions of human transcription factor MEF2C. Brain Res Mol Brain Res 2001; 97:70–82Crossref, Medline, Google Scholar
29 : Postnatal loss of Mef2c results in dissociation of effects on synapse number and learning and memory. Biol Psychiatry 2016; 80:140–148Crossref, Medline, Google Scholar
30 : Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc Natl Acad Sci USA 2008; 105:9397–9402Crossref, Medline, Google Scholar
31 : MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc Natl Acad Sci USA 2008; 105:9391–9396Crossref, Medline, Google Scholar
32 : MEF2C haploinsufficiency syndrome: report of a new MEF2C mutation and review. Eur J Med Genet 2016; 59:478–482Crossref, Medline, Google Scholar
33 : MEF2C deletions and mutations versus duplications: a clinical comparison. Eur J Med Genet 2013; 56:260–265Crossref, Medline, Google Scholar
34 : Genome-wide association meta-analysis of 78,308 individuals identifies new loci and genes influencing human intelligence. Nat Genet 2017; 49:1107–1112Crossref, Medline, Google Scholar
35 : Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511:421–427Crossref, Medline, Google Scholar
36 : Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS Genet 2014; 10:
37 : The transcription factor Mef2 is required for normal circadian behavior in Drosophila. J Neurosci 2010; 30:5855–5865Crossref, Medline, Google Scholar
38 : The transcription factor Mef2 links the Drosophila core clock to Fas2, neuronal morphology, and circadian behavior. Neuron 2013; 79:281–292Crossref, Medline, Google Scholar
39 : Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 2004; 431:862–868Crossref, Medline, Google Scholar
40 : Social experience modifies pheromone expression and mating behavior in male Drosophila melanogaster. Curr Biol 2008; 18:1373–1383Crossref, Medline, Google Scholar
41 : Regulation of feeding and metabolism by neuronal and peripheral clocks in Drosophila. Cell Metab 2008; 8:289–300Crossref, Medline, Google Scholar
42 : Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 2005; 33:D514–D517Crossref, Medline, Google Scholar
43 : A truncating mutation of TRAPPC9 is associated with autosomal-recessive intellectual disability and postnatal microcephaly. Am J Hum Genet 2009; 85:897–902Crossref, Medline, Google Scholar
44 : TRAPPII is required for cleavage furrow ingression and localization of Rab11 in dividing male meiotic cells of Drosophila. J Cell Sci 2009; 122:4526–4534Crossref, Medline, Google Scholar
45 : TRAPPC9-related autosomal recessive intellectual disability: report of a new mutation and clinical phenotype. Eur J Hum Genet 2013; 21:229–232Crossref, Medline, Google Scholar
46 : Combination of linkage mapping and microarray-expression analysis identifies NF-kappaB signaling defect as a cause of autosomal-recessive mental retardation. Am J Hum Genet 2009; 85:903–908Crossref, Medline, Google Scholar
47 : Unlocking the secrets of the genome. Nature 2009; 459:927–930Crossref, Medline, Google Scholar
48 : Imaging analysis of clock neurons reveals light buffers the wake-promoting effect of dopamine. Nat Neurosci 2011; 14:889–895Crossref, Medline, Google Scholar
49 : Circadian rhythm of temperature preference and its neural control in Drosophila. Curr Biol 2012; 22:1851–1857Crossref, Medline, Google Scholar
50 : Sialylation regulates brain structure and function. FASEB J 2015; 29:3040–3053Crossref, Medline, Google Scholar
51 : Molecular mechanism underlying sialic acid as an essential nutrient for brain development and cognition. Adv Nutr 2012; 3(3):465S–472SCrossref, Medline, Google Scholar
52 : Cerebellar neurons lacking complex gangliosides degenerate in the presence of depolarizing levels of potassium. Proc Natl Acad Sci USA 2001; 98:307–312Crossref, Medline, Google Scholar
53 : Genome-wide association study identifies 74 loci associated with educational attainment. Nature 2016; 533:539–542Crossref, Medline, Google Scholar
54 : Epigenetic profiling of ADHD symptoms trajectories: a prospective, methylome-wide study. Mol Psychiatry 2017; 22:250–256Crossref, Medline, Google Scholar
55 : Evidence for selective attention in the insect brain. Curr Opin Insect Sci 2016; 15:9–15Crossref, Medline, Google Scholar



