Temporal, Diagnostic, and Tissue-Specific Regulation of NRG3 Isoform Expression in Human Brain Development and Affective Disorders
Abstract
Objective:
Genes implicated in schizophrenia are enriched in networks differentially regulated during human CNS development. Neuregulin 3 (NRG3), a brain-enriched neurotrophin, undergoes alternative splicing and is implicated in several neurological disorders with developmental origins. Isoform-specific increases in NRG3 are observed in schizophrenia and associated with rs10748842, a NRG3 risk polymorphism, suggesting NRG3 transcriptional dysregulation as a molecular mechanism of risk. The authors quantitatively mapped the temporal trajectories of NRG3 isoforms (classes I–IV) in the neocortex throughout the human lifespan, examined whether tissue-specific regulation of NRG3 occurs in humans, and determined if abnormalities in NRG3 transcriptomics occur in mood disorders and are genetically determined.
Method:
NRG3 isoform classes I–IV were quantified using quantitative real-time polymerase chain reaction in human postmortem dorsolateral prefrontal cortex from 286 nonpsychiatric control individuals, from gestational week 14 to 85 years old, and individuals diagnosed with either bipolar disorder (N=34) or major depressive disorder (N=69). Tissue-specific mapping was investigated in several human tissues. rs10748842 was genotyped in individuals with mood disorders, and association with NRG3 isoform expression examined.
Results:
NRG3 classes displayed individually specific expression trajectories across human neocortical development and aging; classes I, II, and IV were significantly associated with developmental stage. NRG3 class I was increased in bipolar and major depressive disorder, consistent with observations in schizophrenia. NRG3 class II was increased in bipolar disorder, and class III was increased in major depression. The rs10748842 risk genotype predicted elevated class II and III expression, consistent with previous reports in the brain, with tissue-specific analyses suggesting that classes II and III are brain-specific isoforms of NRG3.
Conclusions:
Mapping the temporal expression of genes during human brain development provides vital insight into gene function and identifies critical sensitive periods whereby genetic factors may influence risk for psychiatric disease. Here the authors provide comprehensive insight into the transcriptional landscape of the psychiatric risk gene, NRG3, in human neocortical development and expand on previous findings in schizophrenia to identify increased expression of developmentally and genetically regulated isoforms in the brain of patients with mood disorders. Principally, the finding that NRG3 classes II and III are brain-specific isoforms predicted by rs10748842 risk genotype and are increased in mood disorders further implicates a molecular mechanism of psychiatric risk at the NRG3 locus and identifies a potential developmental role for NRG3 in bipolar disorder and major depression. These observations encourage investigation of the neurobiology of NRG3 isoforms and highlight inhibition of NRG3 signaling as a potential target for psychiatric treatment development.
Genetic studies in humans have identified the neuregulin 3 gene (NRG3) as a candidate risk gene for a range of neurodevelopmental disorders, whereby structural and genetic variations within the gene are associated with developmental delay, attention deficit hyperactivity disorder, bipolar disorder, and schizophrenia (1–11). Emerging evidence also suggests a key role for NRG3 in murine cortical development and human neocortical function (12–14), and recent rodent studies provide in vivo support for a key neurological role of NRG3, with ontogenic models of altered NRG3 signaling displaying abnormal affective behaviors, such as heightened anxiety, increased impulsivity, and reduced social functioning (15–17).
Similar to NRG1, NRG3 undergoes complex alternative splicing and promoter use, generating at least 15 alternative transcripts (7, 13, 18). Our recent cloning of the gene and transcriptional studies in human brain characterized these isoforms into four “classes” based on exon homology, termed NRG3 classes I–IV (7). Interestingly, the NRG3 classes appear to display distinct roles in the human brain, with classes II and IV having significantly higher expression in fetal neocortex compared with postnatal neocortex (7). Despite the emerging evidence for NRG3′s role in neurodevelopment and psychiatric risk, and the potential for the NRG3 classes displaying unique functions, the developmental expression trajectories of NRG3 isoforms in the human brain are unknown.
The NRG3 gene spans 1.2 Mb on human chromosome 10q22-q23 and encodes a transmembrane epidermal growth factor protein belonging to the neuregulin superfamily (19). Linkage with 10q22–q23 and schizophrenia has been confirmed in several populations (2, 3, 5), with subsequent fine mapping identifying a 13-kb interval in intron 1 of NRG3 containing polymorphisms in strong linkage disequilibrium (rs10883866, rs10748842, and rs658440), proxied by rs10748842 (9). Genetic variation at rs10748842 is also associated with behavioral traits in patients, including delusion severity, positive and negative symptom severity, and attentional processes (6–9), as well as prefrontal cortical activation during working memory (12). Moreover, single-nucleotide polymorphisms (SNPs) in NRG3 intron 1 (including rs658440) have recently been shown to be associated with bipolar disorder in a genome-wide association study (GWAS) pathway analysis of psychiatric disorders (20), suggesting that genetic variation at the NRG3 locus may represent a biological mechanism underlying broader psychiatric risk.
While the mechanistic role of rs10748842 remains to be determined, bioinformatics analyses identify the SNP as central to an ultraconserved genomic element and homeodomain binding module that modifies transcription factor binding (7, 18). In the human brain, the risk allele (T) is associated with increased prefrontal cortical expression of NRG3 classes II and III in patients with schizophrenia, nonpsychiatric adult control subjects, and the fetal neocortex, highlighting a potential molecular mechanism of risk (7, 18). Furthermore, classes I and IV are significantly increased in the dorsolateral prefrontal cortex (DLPFC) of schizophrenia patients compared with nonpsychiatric control subjects (7). Collectively, these human studies suggest that balanced regulation of NRG3 transcription plays an important role in normal brain function and that increased NRG3 isoform expression is pathophysiologically relevant to genetic risk for psychiatric disorders.
Given the association of NRG3 with several disorders of neurodevelopment and emergent data suggesting that genes critical to cortical genesis are involved in the pathology of psychiatric illness (20–23), we sought to characterize the temporal trajectories of NRG3 class I–IV expression in the developing human prefrontal cortex, from the second trimester of gestation through adult aging. Furthermore, given the potential genetic overlap between schizophrenia and affective disorders (24) and the recent identification of NRG3-containing GWAS networks associated with bipolar disorder (20), we investigated expression of NRG3 classes I–IV in the DLPFC of patients with bipolar disorder or major depressive disorder and examined the effects of rs10748842 genotype (7). Finally, to determine tissue-specific regulation of NRG3 isoform expression, we compared a comprehensive range of human tissues with brain tissue.
We demonstrate that NRG3 classes I–IV display distinct expression trajectories across neocortical development and aging and that, as in schizophrenia, increased expression of NRG3 is observed in depression and bipolar disorder, in a diagnostic-subclass-specific and genetically regulated manner. Additionally, we present what we believe to be the first tissue-specific mapping of NRG3 class expression in an extensive range of human tissue types, demonstrating that disease-related and genetically associated NRG3 classes II and III are brain-specific. Together, our results provide evidence for differential molecular organization of NRG3 splice isoform expression in human brain development, potentially relevant to the timing of specific neurodevelopmental events, including neuronal migration, cortical patterning, and neural circuit formation. The results also highlight particular splice isoforms in developmental patterns of risk and demonstrate that dysregulation of NRG3 expression occurs in several common psychiatric disorders, further implicating the NRG3 signaling pathway in broader psychiatric risk.
Method
Human Brain Samples
Full details of the postmortem human brain collection used in the current study have been previously described (25). Additional information regarding tissue collection is provided in the data supplement accompanying the online version of this article. A lifespan cohort of 286 nonpsychiatric control subjects ranging in age from gestational week 14 to 85 years old were available for this study. The samples were designated by eight developmental stages based on age at death (26): fetal (<0 year, N=39), neonate (0–0.5 year old, N=19), infant (1–3 years old, N=7), child (4–6 years old, N=7), adolescent (12–18 years old, N=45), young adult (19–25 years old, N=21), adult (26–55 years old, N=110), and aging (56–85 years old, N=38). Full demographic information and information regarding tissue integrity is provided in Table S1 in the online data supplement.
NRG3 expression was examined in mood disorders through inclusion of individuals with a DSM-IV confirmed diagnosis of major depressive disorder (N=69) or bipolar disorder (N=34), compared with an age-matched subgroup (≥18 years old, N=179) of the nonpsychiatric control subjects from the lifespan cohort. Full demographic information, medication status, and information regarding tissue quality and confounding factors are provided in Table S2 in the online data supplement.
RNA Isolation, cDNA Synthesis, and Quantitative Real-Time Polymerase Chain Reaction (PCR)
Tissue homogenates of gray matter dissected from the prefrontal cortex (Brodmann’s area 46/9 in postnatal samples or the corresponding region of the prefrontal cortex in fetal samples) were obtained for all subjects (27). RNA extraction and quantitation were performed as previously described (28). First-strand cDNA was synthesized using the Invitrogen SuperScript First-Strand cDNA Synthesis System as per the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, Mass.). Levels of mRNA expression were measured by means of real-time quantitative PCR using the standard curve method as previously described (7, 28). Expression of NRG3 classes I–IV was assessed using custom designed, and previously validated, exon-spanning TaqMan gene expression assays and normalized to the geometric mean of the mRNA expression of three housekeeping genes—β-actin, HMBS, and GAPDH—within the same sample, as described previously (7). The locations of assay design for NRG3 classes I–IV are shown in Figure S1 in the online data supplement.
For the qualitative comparison of NRG3 expression across neuronal and peripheral tissue types, commercial total RNA derived from pathologically normal human tissues from the hippocampus, prefrontal cortex, breast, heart, kidney, lung, placenta, skeletal muscle, testis, and epidermal fibroblasts was used (Clontech, Takara Bio, Mountain View, Calif.; Ambion, Thermo Fisher Scientific, Waltham, Mass.; and BioChain, Newark, Calif.). Comparison of expression levels between tissues was carried out as in our previous study (29). In brief, tissue samples were run in quadruplicate and NRG3 class expression was normalized to the expression of HMBS (expression of which was similar across tissue types). Normalized expression levels are presented as fold change relative to the expression in the hippocampal sample, using the 2–ΔΔCT method (29, 30).
Genotyping
Genotyping of NRG3 rs10748842 was performed on DNA from patients diagnosed with either bipolar disorder or major depression, as previously described (7). Association of rs10748842 with NRG3 gene expression has been previously reported in the nonpsychiatric control population used here (7). To experimentally verify expression association, we also genotyped rs6584400, a schizophrenia-risk SNP physically located in the 13-kb risk region (7). rs10748842 and rs6584400 are in strong linkage disequilibrium (D′=1, r2=0.85) and likely do not represent independent signals. The genotyping failure rate was less than 1%, and genotyping reproducibility was generally greater than 99%.
Western Blot Analysis
Western blot analysis was performed using standard immunoblot procedures as previously described (7), with the minor modification that immunoreactive bands were visualized using the FluorChem Q Imaging System (Protein Simple, San Jose, Calif.) using an antibody to the N-terminus of human NRG3 (sc-67002, Santa Cruz Biotechnology, Dallas, Tex.) at a dilution of 1:1000. To qualitatively determine the expression of NRG3 protein in neuronal and peripheral tissue types, 35 µg of protein from human cortex, hippocampus, testis, fibroblasts, heart, liver, lung, breast, and skeletal muscle (Clontech, Biochain) was used.
Statistical Analysis
Statistical analyses were performed using SPSS, version 23 (IBM, Armonk, N.Y.). Effects of developmental stage, diagnosis, and genetic variation at rs10748842 (and rs6584400) on NRG3 expression were investigated for each class separately using analysis of covariance (ANCOVA), controlling for appropriate demographic variables and confounding factors. Full details of statistical models used are provided in the supporting text in the online data supplement. The reported degrees of freedom differ slightly across analyses due to either missing gene expression data (absence of detectable expression) or removal of individuals identified as outliers (>2 standard deviations outside of the group mean). The numbers of subjects in the lifespan cohort and in the diagnostic cohort are provided in the online data supplement. All experiments and analyses were conducted by an investigator blind to diagnosis and genotype.
Results
Distinct Developmental Expression Trajectories of NRG3 Classes in Prefrontal Cortex
All NRG3 classes were expressed in the prefrontal cortex across all developmental stages. We observed a significant main effect of developmental stage on NRG3 class I expression (F=5.508, df=7, 269, p=6.19×10−6), with the highest levels of expression during the neonatal period and infanthood (Figure 1A). Expression of NRG3 class I was significantly lower in the fetal brain compared with the neonatal stage and infanthood, and expression was significantly higher during these two postbirth stages than later stages (neonate versus fetus, child, adolescent, young adult, adult, aging: p<0.001; infant versus child, aging: p<0.01; infant versus fetal, adolescent, young adult, adult: p<0.001). By childhood, class I expression had decreased to levels comparable to those observed in the fetus, and it remained stable across the rest of the lifespan.
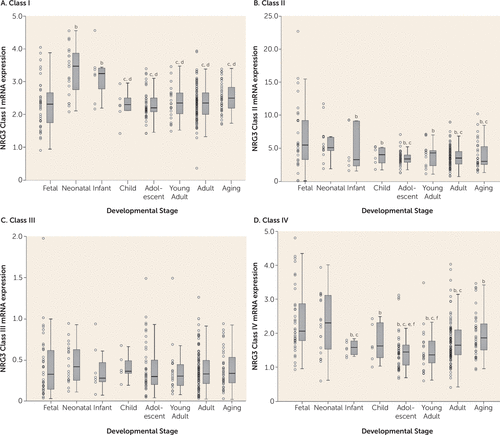
FIGURE 1. NRG3 Class I–IV mRNA Expression in the Dorsolateral Prefrontal Cortex Across the Lifespana
a Fetal=gestational weeks 14–39; neonate=0–0.5 year old; infant=1–3 years old; child=4–6 years old; adolescent=12–18 years old; young adult=19–25 years old; adult=26–55 years old; aging=56–85 years old. Boxplots represent interquartile range above and below the median (horizontal line within box), with whiskers indicating the 1.5× interquartile range. The quantitative, normalized gene expression of each subject is designated by an individual data point.
b p<0.05 compared with fetal stage.
c p<0.05 compared with neonate stage.
d p<0.05 compared with infanthood stage.
e p<0.05 compared with adult stage.
f p<0.05 compared with aging stage.
NRG3 class II expression also was significantly associated with developmental stage (F=3.386, df=7, 216, p=0.002). However, in contrast to class I, NRG3 class II was highest during the fetal and neonatal stages compared with all other developmental stages (fetus versus infant: p<0.05; fetus versus child, adolescent, young adult, adult, aging: p<0.01; neonate versus aging: p<0.05; neonate versus adolescent, adult: p<0.01) (Figure 1B). By infanthood, NRG3 class II expression declined, and it was maintained throughout childhood and into aging.
In contrast, prefrontal cortex NRG3 class III expression was not associated with developmental stage (F=1.846, df=7, 268, p>0.05), with stable levels observed across the lifespan (Figure 1C).
Similar to class II, NRG3 class IV was expressed significantly more during fetal and neonatal periods and decreased in young adulthood, and thus it was strongly associated with developmental stage (F=4.633, df=7, 271, p=6.45×10−5). However, in contrast to expression levels stabilizing across postnatal life, expression of NRG3 class IV began to rise again in adulthood and by old age had reached levels comparably high to those observed during neonatal life (fetus versus child, aging: p<0.05; fetus versus infant: p<0.01; fetus versus adolescent, young adult, adult: p<0.001; adult versus adolescent: p<0.05; aging versus young adult: p<0.01; aging versus adolescent: p<0.001) (Figure 1D). No significant correlations were observed between age and expression of the individual housekeeping genes (GAPDH, β-actin, and HMBS) or between age and the geometric mean used for normalization (r=−0.079–0.099, p>0.18).
Increased NRG3 Class Expression in Mood Disorders
Given previous observations that NRG3 expression is increased in a class-specific manner in the DLPFC of patients with schizophrenia, we examined whether expression changes were evident in other psychiatric disorders, including bipolar disorder and major depressive disorder, to which NRG3 has recently been genetically linked (6, 20). Remarkably, expression levels of NRG3 class I were significantly elevated in patients with bipolar disorder (F=7.771, df=1, 204, p=0.006) and in patients with depression (F=14.126, df=1, 238, p=2.14×10−4) compared with age-matched nonpsychiatric control subjects (Figure 2A), similar to previous findings in schizophrenia (7). In addition, cortical NRG3 class II was significantly increased in bipolar disorder compared with nonpsychiatric controls (F=3.835, df=1, 162, p=0.05) (Figure 2B), and NRG3 class III was significantly elevated in depression (F=12.584, df=1, 238, p=4.67×10−4 (Figure 2C). Expression levels of NRG3 class IV were not changed in bipolar disorder or depression (p>0.05) (Figure 2D). No effects of diagnosis on expression levels of the housekeeping genes used for normalization purposes were observed. Next we investigated the effect of potential confounding variables on NRG3 gene expression in depression and bipolar disorder; we did not observe an association of NRG3 expression with antipsychotic or antidepressant medication status or with suicide as a cause of death (full results available in the supporting text in the online data supplement). In contrast, NRG3 class III expression was associated with nicotine status in all diagnostic groups (full results available in the supporting text in the online data supplement).
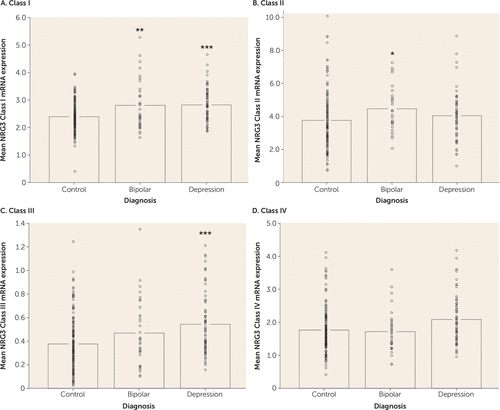
FIGURE 2. NRG3 Class I–IV mRNA Expression in Mood Disordersa
a Nonpsychiatric control individuals were age-matched with individuals diagnosed with bipolar disorder or major depressive disorder. Data bars represent the mean gene expression, with individual data points representing quantitative, normalized subject-level gene expression. Significant differences from the control group are indicated by asterisks.
*p<0.05. ** p<0.01. *** p<0.001.
Association of rs10748842 With NRG3 Class II and III Expression in Depression and Bipolar Disorder
The effect of genetic variation at rs10748842 on expression levels of NRG3 classes previously showing association in patients with schizophrenia and unaffected control subjects (7) was examined in individuals with bipolar disorder or depression. A main effect of genotype at rs10748842 was observed on class II expression (F=12.845, df=1, 83, p=0.001) and class III expression (F=11.371, df=1, 99, p=0.001). No diagnosis-by-genotype interaction was observed (class II and class III, p>0.05), with individuals homozygous for the T allele exhibiting significantly higher NRG3 class II and class III levels compared with individuals heterozygous for the C allele in both bipolar disorder and depression (Figures 3A and 3B). Similarly, a main effect of rs6584400 genotype was confirmed for NRG3 class II (F=13.963, df=1, 82, p=3.41×10−4) and class III (F=7.274, df=1, 99, p=0.008) expression levels in both bipolar disorder and depression (genotype-by-diagnosis interaction, class II and class III, p>0.05) (Figures 3C and 3D). Additionally, since NRG3 class III expression was affected by nicotine status, we examined the impact of nicotine status on NRG3 class III expression with regard to rs10748842 genotype. There was no significant effect of the interaction between nicotine status and genotype at either SNP on NRG3 class III expression (rs10748842 and rs6584400, p>0.05) and no significant interaction of diagnosis, nicotine status, and genotype at either SNP (rs10748842 and rs6584400, p>0.05).
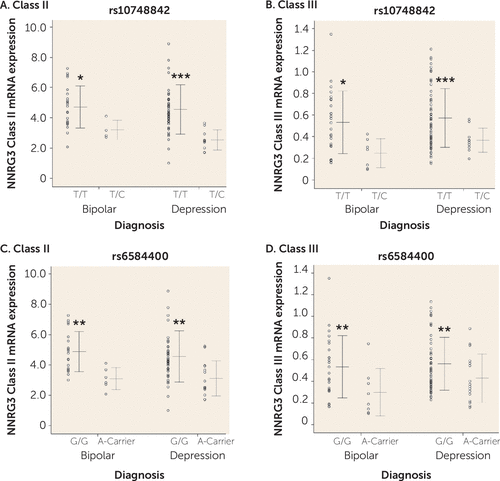
FIGURE 3. Association of NRG3 Class I–IV mRNA Expression With rs10748842 and rs6584400 Genotypes in Mood Disordersa
a Individual data points represent quantitative, normalized subject-level gene expression, with the horizontal line indicating the groups’ mean gene expression and whiskers indicating the standard deviation of the mean. Part A, TT genotype: bipolar disorder, N=22; depression, N=49. Part A, TC genotype: bipolar disorder, N=4; depression, N=10. Part B, TT genotype: bipolar disorder, N=25; depression, N=59. Part B, TC genotype: bipolar disorder, N=7; depression, N=10). Part C, GG genotype: bipolar disorder, N=19; depression, N=45. Part C, A-carrier genotype: bipolar disorder, N=6; depression, N=14. Part D, GG genotype: bipolar disorder, N=24; depression, N=53. Part D, A-carrier genotype: bipolar disorder, N=8; depression, N=16. Significant differences from the nonrisk group are indicated by asterisks.
*p<0.05. ** p<0.01. ***p<0.001.
Tissue Specificity of NRG3 Class Expression
We profiled NRG3 class I–IV mRNA expression in a range of human tissues; including the brain (hippocampus and cortex), breast, heart, kidney, liver, lung, placenta, skeletal muscle, testis, and fibroblasts. NRG3 class I expression was detected in all tissues except fibroblasts. While expression was more abundant in brain tissue than in peripheral tissues, cycle threshold (CT) values of peripheral tissues ranged from 29 to 34, suggesting that NRG3 class I expression is also expressed outside of the brain (Figure 4A). NRG3 classes II and III were detected abundantly in both the adult hippocampus and adult cortex. However, they were not detectable in any peripheral tissues (CT >35) (Figures 4B and 4C), suggesting that these isoforms of NRG3 are brain-specific. NRG3 class IV was also highly expressed in both cortical and hippocampal tissue; however, it was also detectable in the testis, at a slightly lower abundance than that of the brain, and also in the lung, where its expression was approximately threefold lower than that of the hippocampus (Figure 4D). To verify the presence or absence of NRG3 in these tissues, we additionally screened for NRG3 protein in neuronal and peripheral tissues using an N-terminal antibody corresponding to a human NRG3 sequence spanning exons 4 and 5. This antibody is predicted to detect NRG3 classes I, II, and IV (7). In agreement with the mRNA observations, an immunoreactive band of the predicted molecular weight of NRG3 (75 kDa) was detected in all tissues examined except fibroblasts. Interestingly, in contrast to mRNA, protein expression of NRG3 appeared more abundant in peripheral tissues than in the brain, with the highest expression detected in testis, lung, and breast (Figure 4E). The reason for the discordance in tissue mRNA and protein levels is currently unclear.
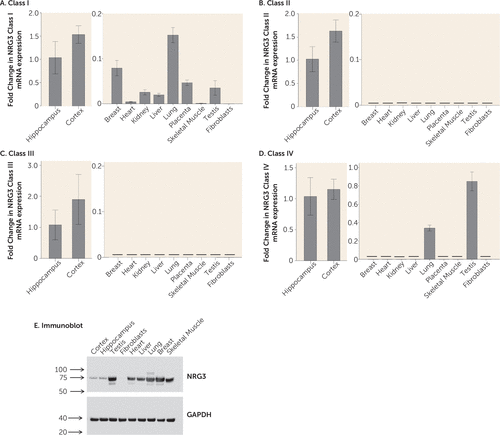
FIGURE 4. NRG3 Class I–IV mRNA Expression in Neuronal and Peripheral Tissuesa
a Parts A–D: Expression in neuronal tissues is shown in the left panel, and expression in peripheral tissues appears in the right panel; note the differing scales of the y axis in the left and right panels. Data represents the fold change in mean normalized mRNA expression relative to expression in the hippocampus (2-ΔΔCT) plus or minus the standard deviation calculated from quadruplicate samples. The horizontal black line denotes no detectable expression in the tissue type, i.e., cycle threshold (CT) >35. CT range for brain expression: class I, 25.8–26.7; class II, 23.7–24.6; class III, 27.1–28.5; class IV, 26.6–27.2. Part E: Immunoblot of neuronal and peripheral tissue protein expression of NRG3 using an N-terminal antibody to human NRG3 (upper panel) and reprobed with an antibody to human GAPDH control (lower panel). Numerical values indicate size in kDa.
Discussion
Structural and polymorphic variation in NRG3 is associated with schizophrenia, bipolar disorder, and several neurodevelopmental disorders characterized by cognitive dysfunction (1–11, 20), implicating NRG3 as a potential key mediator of human neocortical development. We have previously demonstrated that NRG3 undergoes complex differential splicing in human neocortex and that rs10748842, a schizophrenia-risk-associated SNP, is a putative functional polymorphism associated with elevated NRG3 class expression in the brain (7) and altered prefrontal cortical function (12), highlighting potential molecular and neurobiological mechanisms of risk. We now find that NRG3 isoforms, including those modulated by risk-associated genomic variation, exhibit distinct expression trajectories during development and reveal alterations in balanced expression in the DLPFC of patients with mood disorders, as well as schizophrenia (7). This new information provides insights into periods of potential sensitive vulnerability in the immature brain whereby dysregulation of NRG3 expression may contribute to the developmental origins of psychopathology, highlights potentially distinct biological roles for the different NRG3 isoforms, and suggests involvement of NRG3 in the shared biology of three common psychiatric disorders.
During brain development, gene expression patterns are highly dynamic and correlate with the growth and maturation of specific neuroanatomical systems (31–34), and recent molecular genetic studies suggest that variations in genes influencing cortical neurogenesis are involved in the pathogenesis of schizophrenia and related psychiatric disorders (20, 21, 23, 35, 36). Our results provide quantitative molecular evidence that NRG3 isoform classes display unique neuro-ontogenic expression profiles in the human prefrontal cortex, potentially indicative of their key biological roles. As such, we show that NRG3 classes II and IV are dominantly expressed during the prenatal (second trimester) and neonatal period, decreasing to reach adult levels of expression by 1 year of age. Notably, the second trimester of brain development is characterized by major periods of neocortical neuronal migration, organization of the laminar structure of the cortex, and the development of thalamocortical projections (32, 37–39), while the neonatal period is exemplified by the rapid onset of synaptogenesis and oligodendrogenesis (34). These findings emphasize a novel prospective role for NRG3 classes II and IV in human fetal and early postnatal prefrontal cortex development, perhaps consistent with murine studies showing NRG3′s critical contribution to cortical patterning and neuronal migration in the developing cortex (14) and NRG3 class II proteins (hFBNRG3) as oligodendrocyte survival factors (13). Interestingly, several lines of evidence support the centrality of the prefrontal cortex and altered executive function in schizophrenia and bipolar disorder (40–43); therefore it is particularly noteworthy that NRG3 class IV expression is increased in the DLPFC of patients with schizophrenia (7) and class II is increased in bipolar disorder. These findings highlight a potential “immature” pattern of neocortical NRG3 isoform expression in schizophrenia (7) and bipolar disorder and signify a potential developmental and molecular mechanism of risk contributing to altered prefrontal-mediated cognitive function (8, 12).
Consistent with a proposed biological role of NRG3 in human corticogenesis (20, 44) and the suggested developmental origins of psychiatric illness (45, 46), we also present data showing that NRG3 class I expression is significantly increased in bipolar disorder and depression, similar to findings in schizophrenia (7). Interestingly, NRG3 class I isoforms display a unique developmental profile, being >1.5-fold lower in fetal brain, compared with peak expression at 0–3 years of age. This pattern suggests a key role for class I isoforms in postnatal brain development, a time of rapid cortical expansion, synaptogenesis, and progressive myelination (34, 47), and demonstrates that all three common psychiatric disorders share a related molecular perturbation in NRG3. Nevertheless, how this relates to genetic risk is unclear. Although there is evidence of genetic overlap between severe psychiatric disorders, particularly schizophrenia and bipolar disorder, including NRG3 (20, 21), the absence of association of rs10748842 and rs6548440 with class I expression levels suggests a state-related phenomenon. At present it is unclear whether changes in NRG3 class I are epiphenomenal or secondary to changes in other NRG3 isoforms.
Polymorphisms in a 13-kb interval of NRG3 (rs10883866, rs10748842, and rs6584400) are associated with schizophrenia and several core phenotypic features of the illness (7–9, 12). NRG3 rs10748842 resides in a noncoding DNA ultraconserved element and is predicted to be central to a core transcription factor binding sequence for several key developmental transcription factors, including PAX and Hox (7). Biologically, rs10748842 (T allele) is associated with increased expression of NRG3 classes II and III in the DLPFC of control individuals and patients with schizophrenia (7). Our current data provide independent replication of this molecular association in patients with bipolar disorder and depression. Furthermore, we also identify subtle, but significant increases in NRG3 classes II and III in bipolar disorder and depression, respectively, in the same direction as the effects of the risk (T) allele, highlighting a potential molecular etiology underlying genetic risk at NRG3 for affective psychiatric illness and potentially consistent with recent GWAS data identifying NRG3 SNPs within intron 1 (including rs6584400) in the top pathways associated with bipolar disorder (20, 44). As noted above, the developmental pattern of expression of these isoforms is distinct, with class II showing dominant expression in the fetal and neonatal cortex, perhaps consistent with a developmental component to this molecular genetic association, and class III showing maintenance of expression across all developmental stages, suggesting that the genetic association of rs10748842 genotype with class III expression is temporally agnostic. It is also worth noting that class III isoforms represent a potentially nontraditional NRG3 protein due to a truncated epidermal growth factor–like domain (7). Analogous variants with unique signaling properties and cellular distribution have also been identified in the NRG1 family and shown to be relevant to risk for schizophrenia (27). Moreover, tissue expression profiling strongly suggests that NRG3 classes II and III are brain-specific isoforms and we further identify that class III NRG3 expression is significantly associated with smoking status in the human brain. This is particularly interesting given NRG3′s recent genetic association with smoking cessation and nicotine dependence (17, 48) and the increased incidence of nicotine dependence in psychiatric illness.
Finally, while NRG3 expression changes are relatively isoform- and disorder-specific, the directionality of expression changes is consistent with the observation that increased NRG3 signaling is pathologically relevant to psychiatric illness. NRG3′s receptor ErbB4 is also increased in a transcript-dependent manner in schizophrenia (28, 49–51). It is relevant that our recent development of a mouse model of NRG3 overexpression demonstrated that increased NRG3 signaling leads to sustained neurobehavioral deficits in adulthood (16) and inhibition of neuregulin-ErbB4 mediated signaling has shown preclinical utility as a procognitive therapy (52, 53).
In summary, our study provides an important framework for the mechanistic understanding of NRG3’s role in the developmental origins of neuropsychiatric illness and provides novel evidence for NRG3 expression regulation during critical periods of human pre- and postnatal brain development. The findings demonstrate that the NRG3 classes are nonredundant in their temporal, genetic, and tissue-type expression profiles and their involvement in psychiatric disorders and highlight the NRG3 signaling pathway as a potential target for psychiatric drug development.
1 : Recurrent 10q22-q23 deletions: a genomic disorder on 10q associated with cognitive and behavioral abnormalities. Am J Hum Genet 2007; 80:938–947Crossref, Medline, Google Scholar
2 : Genome scan of Han Chinese schizophrenia families from Taiwan: confirmation of linkage to 10q22.3. Am J Psychiatry 2006; 163:1760–1766Link, Google Scholar
3 : Genomewide linkage scan for schizophrenia susceptibility loci among Ashkenazi Jewish families shows evidence of linkage on chromosome 10q22. Am J Hum Genet 2003; 73:601–611Crossref, Medline, Google Scholar
4 : Does parental expressed emotion moderate genetic effects in ADHD? an exploration using a genome wide association scan. Am J Med Genet B Neuropsychiatr Genet 2008; 147B:1359–1368Crossref, Medline, Google Scholar
5 : Elucidating the genetic architecture of familial schizophrenia using rare copy number variant and linkage scans. Proc Natl Acad Sci USA 2009; 106:16746–16751Crossref, Medline, Google Scholar
6 : Neuregulin 3 is associated with attention deficits in schizophrenia and bipolar disorder. Int J Neuropsychopharmacol 2013; 16:549–556Crossref, Medline, Google Scholar
7 : Common genetic variation in neuregulin 3 (NRG3) influences risk for schizophrenia and impacts NRG3 expression in human brain. Proc Natl Acad Sci USA 2010; 107:15619–15624Crossref, Medline, Google Scholar
8 : Neuregulin 3 (NRG3) as a susceptibility gene in a schizophrenia subtype with florid delusions and relatively spared cognition. Mol Psychiatry 2011; 16:860–866Crossref, Medline, Google Scholar
9 : Fine mapping on chromosome 10q22-q23 implicates Neuregulin 3 in schizophrenia. Am J Hum Genet 2009; 84:21–34Crossref, Medline, Google Scholar
10 : NRG3 gene is associated with the risk and age at onset of Alzheimer disease. J Neural Transm (Vienna) 2014; 121:183–192Crossref, Medline, Google Scholar
11 : Neuregulin 3 genetic variations and susceptibility to schizophrenia in a Chinese population. Biol Psychiatry 2008; 64:1093–1096Crossref, Medline, Google Scholar
12 : Effects of neuregulin 3 genotype on human prefrontal cortex physiology. J Neurosci 2014; 34:1051–1056Crossref, Medline, Google Scholar
13 : Characterization of a neural-specific splicing form of the human neuregulin 3 gene involved in oligodendrocyte survival. J Cell Sci 2006; 119:898–909Crossref, Medline, Google Scholar
14 : Identification of a Pax6-dependent epidermal growth factor family signaling source at the lateral edge of the embryonic cerebral cortex. J Neurosci 2003; 23:6399–6403Crossref, Medline, Google Scholar
15 : Neuregulin-3 in the mouse medial prefrontal cortex regulates impulsive action. Biol Psychiatry 2014; 76:648–655Crossref, Medline, Google Scholar
16 : Transient overexposure of neuregulin 3 during early postnatal development impacts selective behaviors in adulthood. PLoS One 2014; 9:e104172Crossref, Medline, Google Scholar
17 : Evidence from mouse and man for a role of neuregulin 3 in nicotine dependence. Mol Psychiatry 2014; 19:801–810Crossref, Medline, Google Scholar
18 : Identification and functional studies of regulatory variants responsible for the association of NRG3 with a delusion phenotype in schizophrenia. Mol Neuropsychiatry 2015; 1:36–46Crossref, Medline, Google Scholar
19 : Neuregulin-3 (NRG3): a novel neural tissue-enriched protein that binds and activates ErbB4. Proc Natl Acad Sci USA 1997; 94:9562–9567Crossref, Medline, Google Scholar
20 : Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci 2015; 18:199–209Crossref, Medline, Google Scholar
21 : Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell 2013; 154:518–529Crossref, Medline, Google Scholar
22 : Prenatal expression patterns of genes associated with neuropsychiatric disorders. Am J Psychiatry 2014; 171:758–767Link, Google Scholar
23 : De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet 2012; 44:1365–1369Crossref, Medline, Google Scholar
24 : Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 2013; 45:984–994Crossref, Medline, Google Scholar
25 : Neurexin 1 (NRXN1) splice isoform expression during human neocortical development and aging. Mol Psychiatry 2016; 21:701–706.Crossref, Medline, Google Scholar
26 : Changes in NMDA receptor subunit mRNAs and cyclophilin mRNA during development of the human hippocampus. Ann N Y Acad Sci 2003; 1003:426–430Crossref, Medline, Google Scholar
27 : Effects of schizophrenia risk variation in the NRG1 gene on NRG1-IV splicing during fetal and early postnatal human neocortical development. Am J Psychiatry 2014; 171:979–989Link, Google Scholar
28 : Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum Mol Genet 2007; 16:129–141Crossref, Medline, Google Scholar
29 : Molecular cloning of a brain-specific, developmentally regulated neuregulin 1 (NRG1) isoform and identification of a functional promoter variant associated with schizophrenia. J Biol Chem 2007; 282:24343–24351Crossref, Medline, Google Scholar
30 : Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 2001; 25:402–408Crossref, Medline, Google Scholar
31 : Transcriptional profiling of the developing rat brain reveals that the most dramatic regional differentiation in gene expression occurs postpartum. J Neurosci 2006; 26:345–353Crossref, Medline, Google Scholar
32 : Normal development of brain circuits. Neuropsychopharmacology 2010; 35:147–168Crossref, Medline, Google Scholar
33 : Developmental regulation of human cortex transcription and its clinical relevance at single base resolution. Nat Neurosci 2015; 18:154–161Crossref, Medline, Google Scholar
34 : The cellular and molecular landscapes of the developing human central nervous system. Neuron 2016; 89:248–268Crossref, Medline, Google Scholar
35 : Investigation of the prenatal expression patterns of 108 schizophrenia-associated genetic loci. Biol Psychiatry 2015; 77:e43–e51Crossref, Medline, Google Scholar
36 : Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat Neurosci 2016; 19:40–47Crossref, Medline, Google Scholar
37 : Ontogeny of the human central nervous system: what is happening when? Early Hum Dev 2006; 82:257–266Crossref, Medline, Google Scholar
38 : Structural and functional maturation of the developing primate brain. J Pediatr 2003; 143(Suppl):S35–S45Crossref, Medline, Google Scholar
39 : Transient cholinesterase staining in the mediodorsal nucleus of the thalamus and its connections in the developing human and monkey brain. J Comp Neurol 1983; 219:431–447Crossref, Medline, Google Scholar
40 : Cognitive endophenotypes of bipolar disorder: a meta-analysis of neuropsychological deficits in euthymic patients and their first-degree relatives. J Affect Disord 2009; 113:1–20Crossref, Medline, Google Scholar
41 : Altered prefrontal cortex activity during working memory task in bipolar disorder: a functional magnetic resonance imaging study in euthymic bipolar I and II patients. J Affect Disord 2015; 184:116–122Crossref, Medline, Google Scholar
42 : Complexity of prefrontal cortical dysfunction in schizophrenia: more than up or down. Am J Psychiatry 2003; 160:2209–2215Link, Google Scholar
43 : Impaired intellect and memory: a missing link between genetic risk and schizophrenia? Arch Gen Psychiatry 2010; 67:905–913Crossref, Medline, Google Scholar
44 : Genome-wide association study of bipolar disorder in Canadian and UK populations corroborates disease loci including SYNE1 and CSMD1. BMC Med Genet 2014; 15:2Crossref, Medline, Google Scholar
45 : Neurodevelopmental basis of bipolar disorder: a critical appraisal. Prog Neuropsychopharmacol Biol Psychiatry 2008; 32:1617–1627Crossref, Medline, Google Scholar
46 : Neurodevelopmental origins of depressive disorders. Curr Opin Pharmacol 2007; 7:8–17Crossref, Medline, Google Scholar
47 : Longitudinal development of cortical and subcortical gray matter from birth to 2 years. Cereb Cortex 2012; 22:2478–2485Crossref, Medline, Google Scholar
48 : Role of the neuregulin signaling pathway in nicotine dependence and co-morbid disorders. Int Rev Neurobiol 2015; 124:113–131Crossref, Medline, Google Scholar
49 : The involvement of ErbB4 with schizophrenia: association and expression studies. Am J Med Genet B Neuropsychiatr Genet 2006; 141B:142–148Crossref, Medline, Google Scholar
50 : Elevated ErbB4 mRNA is related to interneuron deficit in prefrontal cortex in schizophrenia. J Psychiatr Res 2014; 53:125–132Crossref, Medline, Google Scholar
51 : Dysregulated ErbB4 splicing in schizophrenia: selective effects on parvalbumin expression. Am J Psychiatry 2016; 173:60–68Link, Google Scholar
52 : Neuregulin 1-ErbB4-PI3K signaling in schizophrenia and phosphoinositide 3-kinase-p110δ inhibition as a potential therapeutic strategy. Proc Natl Acad Sci USA 2012; 109:12165–12170Crossref, Medline, Google Scholar
53 : Behavioral, neurophysiological, and synaptic impairment in a transgenic neuregulin1 (NRG1-IV) murine schizophrenia model. J Neurosci 2016; 36:4859–4875Crossref, Medline, Google Scholar



