Alterations in Metabotropic Glutamate Receptor 1α and Regulator of G Protein Signaling 4 in the Prefrontal Cortex in Schizophrenia
Abstract
Objective:
Certain cognitive deficits in individuals with schizophrenia have been linked to disturbed gamma-aminobutyric acid (GABA) and glutamate neurotrans-mission in the prefrontal cortex. Thus, it is important to understand how the mechanisms that regulate GABA and glutamate neurotransmission are altered in schizophrenia. For example, group I metabo-tropic glutamate receptors (mGluR1α, mGluR5) modulate both GABA and gluta-mate systems. In addition, regulator of G protein signaling 4 (RGS4) reduces intra-cellular signaling through several different G protein-coupled receptors, including group I mGluRs. Finally, the endocannabinoid system plays an important role in regulating GABA and glutamate neurotrans-mission. The status of endocannabinoid ligands, such as 2-arachidonoylglycerol, can be inferred in part through measures of diacylglycerol lipase and monoglyceride lipase, which synthesize and degrade 2-arachidonoylglycerol, respectively.
Method:
Quantitative polymerase chain reaction was used to measure mRNA levels for group I mGluRs, RGS4, and markers of the endocannabinoid system in the prefrontal cortex Brodmann's area 9 of 42 schizophrenia subjects and matched normal comparison subjects. Similar analyses in monkeys chronically exposed to haloperidol, olanzapine, or placebo were also conducted.
Results:
Schizophrenia subjects had higher mRNA levels for mGluR1α and lower mRNA levels for RGS4, and these differences did not appear to be attributable to antipsychotic medications or other potential confounds. In contrast, no differences between subject groups were found in mRNA levels for endocannabinoid synthesizing and metabolizing enzymes.
Conclusions:
Together, higher mGluR1α and lower RGS4 mRNA levels may represent a disturbed “molecular hub” in schizophrenia that may disrupt the function of prefrontal cortical networks, including both GABA and glutamate systems.
Cognitive impairments, such as deficits in working memory, are among the most disabling and difficult to treat features of schizophrenia (1, 2) and appear to be linked to impairments in the circuitry of the prefrontal cortex (3). For example, markers of gamma-aminobutyric acid (GABA) neurotransmission are altered in subpopulations of prefrontal cortex GABA neurons (4), and evidence of disturbed glutamate neurotransmission, such as N-methyl-D-aspartic acid (NMDA) receptor hypo-function (5), has also been reported in the illness. Thus, understanding the mechanisms that might contribute to disturbed GABA and glutamate neurotransmission is essential for developing novel treatments for cognitive impairments in schizophrenia.
Group I metabotropic glutamate receptors (mGluR1α, mGluR5) play an important role in regulating GABA and glutamate neurotransmission in the cortex. For example, activation of group I mGluRs initiates an intracellular signaling cascade that leads to suppression of GABA release (6–10) and long-term potentiation of NMDA receptor function (11). Interestingly, one study reported higher protein levels of mGluR1α, but not mGluR5, in the prefrontal cortex of elderly schizophrenia subjects (12), which may contribute to altered GABA and glutamate neurotrans-mission in schizophrenia. However, it remains unclear whether group I mGluR alterations are generalizable to most subjects with schizophrenia, including younger individuals with the disorder, and whether alterations in group I mGluR protein levels are attributable to differences in gene expression levels.
Activation of group I mGluRs initiates a G protein signaling pathway, which is inhibited by regulator of G protein signaling 4 (RGS4) (13). RGS4 functions as a GTPase-activating protein that increases the hydrolysis of guanosine triphosphate (GTP) and reduces the duration of activity of several different G protein-coupled receptors, including group I mGluRs (13). RGS4 has been previously identified as a gene of interest in schizophrenia because of genetic association studies (14) and reports of lower expression levels in the prefrontal cortex in two relatively small cohorts of subjects with schizophrenia (15, 16). However, it is unclear whether lower RGS4 mRNA levels are commonly found in schizophrenia and whether these lower levels, which may affect the duration of intracellular signaling from group I mGluRs, are present in the same schizophrenia subjects who have higher group I mGluR levels.
The endogenous cannabinoid system is also critically involved in regulating GABA and glutamate neurotrans-mission in the cortex. For example, the endocannabinoid 2-arachidonoylglycerol is synthesized by diacylglycerol lipase (17) in pyramidal neurons (18, 19) and then travels retrogradely to stimulate cannabinoid 1 receptors located in nearby axon terminals from certain inhibitory neurons (20) and, to a lesser extent, other pyramidal neurons (21), leading to a suppression of GABA and gluta-mate release (21, 22). 2-Arachidonoylglycerol activity is then terminated through metabolism by monoglyceride lipase (23). Interestingly, the endocannabinoid system also appears to be the primary mediator of the aforementioned suppressive effects of group I mGluRs on GABA release. For example, activation of group I mGluRs, but not other classes of mGluRs, leads to increased synthesis of 2-arachidonoylglycerol (24) and suppression of GABA release (6–10) in a manner that is dependent upon the cannabinoid 1 receptor (7, 9, 10). We recently reported lower cannabinoid 1 receptor mRNA and protein levels in the prefrontal cortex in individuals with schizophrenia (25). However, fully understanding the pathophysiological state of the endocannabinoid system as well as the downstream effects on GABA and glutamate neuro-transmission in schizophrenia also requires knowledge of whether the level of 2-arachidonoylglycerol is altered in the disorder. Levels of 2-arachidonoylglycerol cannot be directly assessed in postmortem human brain tissue (26) but can be inferred, in part, by measures of gene products that regulate 2-arachidonoylglycerol synthesis (diacylglycerol lipase) and degradation (monoglyceride lipase). To our knowledge, no studies have yet reported the status of endocannabinoid ligands in schizophrenia.
Therefore, in the present study, we sought to evaluate the status of several important regulators of GABA and glutamate neurotransmission in the prefrontal cortex by examining the transcript levels of mGluR1α, mGluR5, RGS4, and endocannabinoid synthesizing (diacylglycerol lipase) and metabolizing (monoglyceride lipase) enzymes in a relatively large cohort of schizophrenia subjects (N=42). In schizophrenia subjects, we found elevated mRNA levels for mGluR1α, but not mGluR5, and lower RGS4 mRNA levels, but transcript levels for diacylglycerol and monoglyceride lipase did not differ between subject groups. Higher mGluR1α and lower RGS4 mRNA levels may have an important impact on GABA and glutamate neurotransmission in schizophrenia.
Method
Human Subjects
Brain specimens were obtained during routine autopsies conducted at the Allegheny County Medical Examiner's Office (Pittsburgh) after consent was obtained from the next-of-kin. An independent committee of experienced research clinicians made consensus DSM-IV diagnoses for each subject using the results of structured interviews conducted with family members and review of medical records, as previously described (27). In order to control for experimental variance, subjects with schizophrenia or schizoaffective disorder (N=42) were matched individually with normal comparison subjects for sex and as closely as possible for age and postmortem interval (Table 1 [also see Table S1 in the data supplement accompanying the online version of this article for demographic details on individual subjects]), and samples from both subjects in a pair were processed together throughout all stages of the study. The mean age, postmortem interval, brain pH, RNA integrity number (as determined from Agilent Bioanalyzer 2100, Agilent Technologies, Walbronn, Germany [4, 28]), and tissue storage time did not differ between the subject groups. All procedures were approved by the University of Pittsburgh Committee for the Oversight of Research Involving the Dead and Institutional Review Board for Biomedical Research.
| Characteristic | Comparison Subjects (N=42) | Schizophrenia Subjects (N=42) | ||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| Age (years) | 48 | 13 | 47 | 13 |
| Postmortem interval (hours) | 17.8 | 5.9 | 18.1 | 8.7 |
| Freezer storage time (months) | 97 | 43 | 97 | 46 |
| Brain pH | 6.8 | 0.2 | 6.6 | 0.4 |
| RNA integrity number | 8.3 | 0.6 | 8.2 | 0.7 |
| N | % | N | % | |
| Sex | ||||
| Male | 31 | 73.8 | 31 | 73.8 |
| Female | 11 | 26.2 | 11 | 26.2 |
| Race | ||||
| Caucasian | 34 | 80.9 | 29 | 69.0 |
| African American | 8 | 19.0 | 13 | 30.9 |
| Diagnosis | ||||
| Schizophrenia | 28 | 66.7 | ||
| Schizoaffective disorder | 14 | 33.3 | ||
| Suicide | 11 | 26.2 | ||
| Substance disorder diagnosisb | 13 | 31.0 | ||
| Antidepressant useb | 17 | 40.5 | ||
| Benzodiazepine/valproate useb | 15 | 35.7 | ||
| Antipsychotic useb | 36 | 85.7 | ||
| History of tetrahydrocannabinol use | 14 | 33.3 | ||
TABLE 1. Demographic, Clinical, and Postmortem Characteristics of Schizophrenia Subjects and Matched Normal Comparison Subjectsa
Tissue Preparation
The right hemisphere of each brain was blocked coronally, immediately frozen, and stored at −80°C (29). Cryostat sections, from the anterior-posterior level corresponding to the middle portion of the superior frontal sulcus, were cut serially and confirmed to contain prefrontal cortex Brodmann's area 9, from adjacent Nissl-stained sections using cytoarchitectonic criteria (29). The cortical gray matter was dissected in a manner that excluded white matter contamination and provided excellent RNA preservation, and the gray matter was collected into tubes containing Trizol reagent (Invitrogen, Carlsbad, Calif.) for RNA isolation (30). Total RNA was isolated from Trizol homogenates and cleaned using RNeasy columns (Qiagen, Valencia, Calif.).
Real-Time Reverse Transcription PCR
For each subject, cDNA was synthesized from total RNA using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, Calif.). All primer pairs (see Table S2 in the data supplement) demonstrated 1) high amplification efficiency (>96%) across a wide range of cDNA dilutions; 2) specific single products in dissociation curve analysis; and 3) melting temperatures similar to those predicted by oligonucleotide software. Quantitative polymerase chain reaction (PCR) was performed with the comparative threshold cycle (Ct) measurement using Power SYBR Green dye and StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, Calif.). Based on their stable level of expression between schizophrenia and normal comparison subjects (30), three reference genes (beta actin, cyclophilin A, and glyceraldehyde-3-phosphate dehydrogenase) were used to normalize the target gene expression levels. The difference in cycle threshold for each target transcript was calculated by subtracting the mean cycle threshold for the three reference genes from the cycle threshold of the target transcript. Because this difference in cycle threshold (dCT) represents the log2-transformed expression ratio of each target transcript to the geometric mean of the three reference genes, the relative expression level of the target transcript is determined as 2-dCT (30, 31). Four replicate measures were performed for each gene of interest for each subject, with a standard threshold consistently applied for each gene across all subjects. The following transcripts were assessed (with the mean coefficient of variance in parentheses): mGluR1α (mean: 0.043 [SD=0.023]), mGluR5 (mean: 0.028 [SD=0.011]), RGS4 (mean: 0.025 [SD=0.018]), diacylglycerol lipase isoforms α (mean: 0.035 [SD=0.017]) and β (mean: 0.046 [SD=0.028]) (32), monoglyceride lipase (mean: 0.026 [SD=0.013]), fa tty acid amide hydrolase (mean: 0.037 [SD=0.02]), and cannabinoid receptor interacting protein 1a (mean: 0.030 [SD=0.015]) (see Table S2 in the data supplement). In addition, statistically significant findings were validated by repeating the study in a subset of subject pairs using additional primer sets for nonoverlapping regions of the same targets.
Cannabinoid 1 receptor mRNA and protein levels of the first 23 subjects with schizophrenia and RGS4 mRNA levels of the first eight subjects with schizophrenia included in this study (see Table S1 in the data supplement) were previously shown to be reduced (15, 25).
Antipsychotic-Exposed Monkeys
As described previously (33), experimentally naive, young adult male macaque monkeys (Macaca fascicularis) were administered oral doses of haloperidol, olanzapine, or placebo (monkeys per group: N=6) twice daily for 17–27 months. The final trough drug plasma levels were within the range associated with clinical efficacy in humans (haloperidol: approximately 1.5 ng/ml; olanza-pine: approximately 15 ng/ml) (33). Monkeys were matched by terminal body weight and euthanized in triads (one animal from each group) on the same day. Brains were rapidly removed, and the right frontal lobe was cut into coronal blocks, frozen, and stored at −80°C. RNA was isolated from prefrontal cortex Brod-mann's area 9, and quantitative PCR was conducted for the same three reference genes and mGluR1α (see Table S2 in the data supplement), with all monkeys from a triad processed together on the same plate. Primers were designed using monkey-specific cDNA sequences, and primer amplification efficiencies were >96% in monkey brain tissue. All animal studies were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
Statistical Analysis
Two analysis of covariance (ANCOVA) models (34) were performed to test the effect of diagnosis on relative expression level for each target mRNA (25, 30). The first model included relative expression level as the dependent variable; diagnostic group as the main effect; storage time, brain pH, and RNA integrity number as covariates; and subject pair as a blocking factor. Subject pairing may be considered an attempt to balance diagnostic groups for sex, age, and postmortem interval and to account for the parallel processing of tissue samples from a pair and not a true statistical paired design. Therefore, we also utilized a second model without subject pair as a blocking factor that included all pairing factors (age, sex, and postmortem interval), storage time, brain pH, and RNA integrity number as covariates. Because these two ANCOVAs produced nearly identical results, only the results from the first model are reported. All statistical tests are two-tailed. The Bonferroni correction for multiple comparisons was employed to set the statistical significance for the seven newly studied target mRNAs in this cohort to an alpha of 0.007.
The effects of potential confounding variables on relative mRNA expression levels in subjects with schizophrenia were assessed using an ANCOVA model with each potential confounding variable as the main effect and with sex, age, postmortem interval, storage time, pH, and RNA integrity number as covariates. An ANOVA model with relative mRNA expression level as the dependent variable, treatment group as the main effect, and triad as a blocking factor was used for the antipsychotic-exposed monkey study.
Results
Group I mGluRs in Schizophrenia
We found elevated mRNA levels for mGluR1α ( +12.4%; F=9.2, df=1, 38, p=0.004), but not mGluR5, in subjects with schizophrenia (Figure 1). Exclusion of the two subject pairs with the greatest mGluR1α mRNA levels in the schizophrenia group still resulted in significantly higher mGluR1α mRNA levels in schizophrenia subjects (+10.2%; F=7.77, df=1, 36, p=0.008). The relative mRNA expression level of mGluR1α in schizophrenia subjects did not differ as a function of sex; suicide; diagnosis of substance abuse or dependence current at the time of death; the use of antidepressants, benzodiazepines or sodium valproate, or antipsychotics at the time of death; or a history of cannabis use (all F≤1.24, p≥0.273) (Figure 2). However, schizophrenia subjects (N=28) may have a higher level (+11.5%) of mGluR1α mRNA expression than schizoaffective disorder subjects (N=14). Furthermore, when the schizophrenia cohort was subdivided into schizophrenia and schizoaffective disorder subjects, schizophrenia subjects had a significantly higher level of mGluR1α mRNA expression compared with their matched normal comparison counterparts (+16.3%; F=9.7, df=1, 24, p=0.005), while schizoaffective disorder subjects did not (+5.4%; F=1.2, df=1, 10, p=0.30). Similarly, but to a lesser extent, schizophrenia subjects may also have a higher level of mGluR5 mRNA expression than schizoaffective disorder subjects (+5.9%; F=5.31, df=1, 35, p=0.027). However, after excluding schizoaffective disorder subjects, mGluR5 mRNA levels still did not differ between schizophrenia and matched normal comparison subjects (+5.1%; F=2.49, df=1, 24, p=0.127). Finally, higher mGluR1α mRNA expression levels in the prefrontal cortex in schizophrenia subjects were validated in a subset of subject pairs using additional quantitative PCR primer sets designed against nonover-lapping regions of mGluR1α cDNA (see Figure S1 and Table S2 in the data supplement).
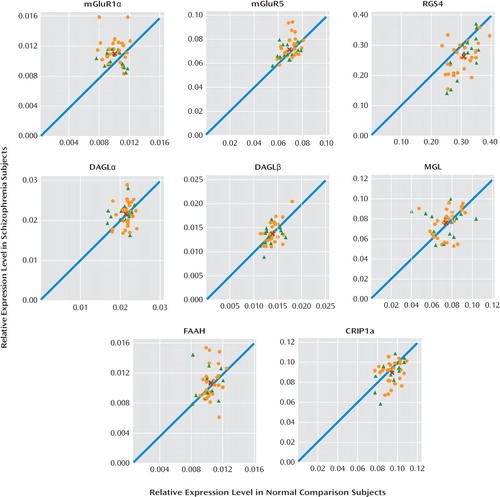
FIGURE 1. Relative Expression Levels of Transcripts in Schizophrenia Subjects Relative to Matched Normal Comparison Subjectsa
a The scatter plots indicate mRNA expression levels for schizophrenia subjects relative to matched normal comparison subjects (orange circles) and mRNA expression levels for schizoaffective disorder subjects relative to matched normal comparison subjects (green triangles). Data points located to the left of the unity line indicate higher levels of mRNA expression in the affected subject relative to the normal comparison subject and vice versa. The average mRNA expression levels for both diagnostic groups are indicated by the red “X” symbol. Statistically significant differences (Bonferroni correction: α=0.007) in mRNA expression levels in schizophrenia subjects were found for mGluR1α (+12.4%; F=9.2, df=1, 38, p=0.004) and RGS4 (−16.1%; F=29.4, df=1, 38, p=0.000). Abbreviations: mGluR=metabotropic glutamate receptor; RGS4=regulator of G protein signaling 4; DAGL=diacylglycerol lipase; MGL=monoglyceride lipase; FAAH=fatty acid amide hydrolase; CRIP1a=cannabinoid receptor interacting protein 1a.
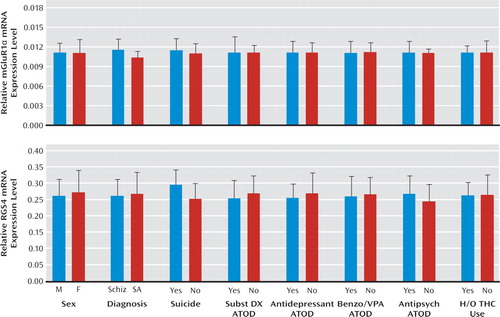
FIGURE 2. Effects of Potentially Confounding Variables on mGluR1α and RGS4 mRNA Expression Levels in Schizophrenia Subjectsa
a Bar graphs indicate that sex, suicide, diagnosis of substance abuse or dependence current at the time of death, antidepressant medication use at the time of death, use of benzodiazepines or sodium valproate at the time of death, antipsychotic medication use at the time of death, and history of cannabis use (tetrahydrocannabinol) did not significantly affect metabotropic glutamate receptor 1α (mGluR1α) mRNA expression levels or regulator of G protein signaling 4 (RGS4) mRNA expression levels. However, schizophrenia subjects may have a higher level of mGluR1α mRNA expression levels than schizoaffective disorder subjects (+11.5%; F=4.42, df=1, 34, p=0.04), with no difference in RGS4 mRNA expression levels. Abbreviations: M=male; F=female; Schiz=schizophrenia subjects; SA=schizoaffective disorder subjects; Subst DX=substance disorder diagnosis; Benzo/VPA=benzodiazepines or valproate use; Antipsych=antipsychotic use; ATOD=at time of death; H/O THC=history of tetrahydrocannabinol.
RGS4 in Schizophrenia
We also examined mRNA levels for RGS4, which have previously been reported to be reduced in the prefrontal cortex in individuals with schizophrenia (15, 16), including in the first eight subjects (15) of the current cohort of 42 schizophrenia subjects (see Table S1 in the data supplement). We found lower RGS4 mRNA expression levels in our entire cohort of schizophrenia subjects relative to matched normal comparison subjects (−16.1%; F=29.4, df=1, 38, p=0.000) (Figure 1) as well as in the previously untested 34 subject pairs alone (−16.3%; F=22.0, df=1, 30, p=0.000). The relative mRNA expression level of RGS4 in schizophrenia subjects did not differ as a function of sex; diagnosis of schizophrenia versus schizoaffective disorder; suicide; diagnosis of substance abuse or dependence current at the time of death; the use of antidepressants, benzodiazepines or sodium valproate, or antipsychotics at the time of death; or a history of cannabis use (Figure 2).
RGS4 reduces the duration of intracellular signaling from group I mGluRs (13). Therefore, we further examined the relationship between RGS4 and mGluR1α mRNA levels in this cohort. Pearson correlation analysis revealed that RGS4 and mGluR1α mRNA levels were not correlated in individual subjects, and differences in RGS4 and mGluR1α mRNA levels between schizophrenia and matched comparison subjects were not correlated in subject pairs. However, a significant number of subject pairs (27 out of 42 pairs) showed both higher mGluR1α and lower RGS4 mRNA levels in the schizophrenia subject relative to the matched normal comparison subject (χ2=37.6, df=3, p=0.000). In contrast, only five subject pairs showed higher levels of both mGluR1α and RGS4 mRNA in the schizophrenia subject relative to the matched normal comparison subject, nine subject pairs showed lower levels of both mGluR1α and RGS4 mRNA in the schizophrenia subject relative to the matched normal comparison subject, and one subject pair showed lower levels of mGluR1α and higher levels of RGS4 mRNA in the schizophrenia subject relative to the matched normal comparison subject.
Synthesizing and Metabolizing Enzymes for Endocannabinoids in Schizophrenia
We quantified mRNA expression levels for both isoforms of the synthesizing enzyme for 2-arachidonoylglycerol (diacylglycerol lipase α and diacylglycerol lipase β), the metabolizing enzyme for 2-arachidonoylglycerol (monoglyceride lipase), and fatty acid amide hydrolase, which is the metabolizing enzyme for another cortical endocannabinoid (anandamide). Relative mRNA expression levels for diacylglycerol lipase α, diacylglycerol lipase β, monoglyceride lipase, and fatty acid amide hydrolase did not differ between the subject groups (all transcripts: maximum difference in group means ≤ 3%; F≤1.1, df=1, 38, p≥0.30) (Figure 1). We also examined mRNA levels for cannabi noid receptor interacting protein 1a, which has been reported to bind to and inhibit the functioning of cannabinoid 1 receptor (35). We found a small difference in cannabinoid receptor interacting protein 1a mRNA expression levels in schizophrenia subjects (−5%; F=5.7, df=1, 38, p=0.022) that was not statistically significant after correction for multiple comparisons (Bonferroni correction: α=0.007; see Statistical Analysis) (Figure 1). In contrast to group I mGluRs, transcript levels for diacylglycerol lipase α, diacylglycerol lipase β, monoglyceride lipase, fatty acid amide hydrolase, and cannabinoid receptor interacting protein 1a did not differ between schizophrenia and schizoaffective disorder subjects (F≤1.37, df=1, 35, p≥0.25).
Antipsychotic-Exposed Monkeys
Prior studies in antipsychotic-exposed monkeys have provided evidence suggesting that lower RGS4 mRNA levels in the prefrontal cortex in schizophrenia do not appear to be attributable to antipsychotic treatment (15). However, the effects of antipsychotic treatment on mGluR1α in the prefrontal cortex have not been established. In order to further assess whether alterations in mGluR1α mRNA expression in schizophrenia are confounded by treatment with antipsychotic medications, we used quantitative PCR to determine relative mRNA expression levels of mGluR1α in monkeys chronically exposed to haloperidol, olanzapine, or placebo. In contrast to our findings in schizophrenia subjects, the relative expression level of mGluR1α mRNA was slightly lower in haloperidol-exposed (−7.7%) and olanzapine-exposed monkeys (−4.7%) compared with placebo-exposed monkeys (Figure 3). However, these differences did not achieve statistical significance (F=0.53, df=2, 15, p=0.60). When the haloperidol- and olanzapine-exposed monkeys were combined into a single group, the relative expression level of mGluR1α mRNA was still slightly, but not statistically significantly, lower in the antipsychotic-exposed monkeys compared with placebo-exposed monkeys (−6.2%; F=0.83, df=1, 5, p=0.41).
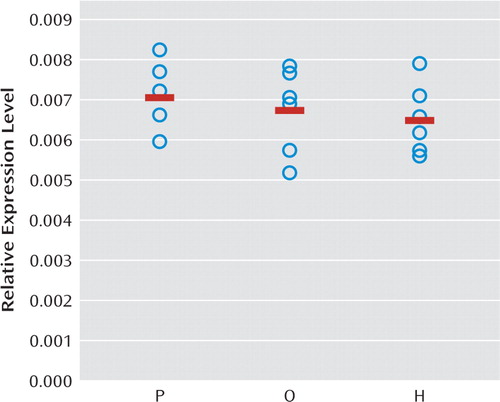
FIGURE 3. Relative mRNA Expression Levels for mGluR1α in Antipsychotic Medication-Exposed Monkeysa
a No statistically significant differences were found in mRNA expression for metabotropic glutamate receptor 1α (mGluR1α) in monkeys chronically exposed to olanzapine (O) or haloperidol (H) relative to placebo (P).
Discussion
In this study, we found that mRNA expression levels for mGluR1α, but not mGluR5, were significantly higher in the prefrontal cortex in schizophrenia subjects. Consistent with prior reports (15, 16), we also found lower mRNA levels for RGS4, which reduces the duration of G protein-mediated intracellular signaling from mGluR1α activation (13). Lower RGS4 mRNA levels were commonly found in the same schizophrenia subjects who had higher mGluR1α mRNA levels. In contrast, no differences were found in mRNA levels for the synthesizing and metabolizing enzymes for 2-arachidonoylglycerol, diacylglycerol lipase (both α and β isoforms), and monoglyceride lipase, respectively, or for fatty acid amide hydrolase, the metabolizing enzyme of the other cortical endocannabinoid, anandamide, in schizophrenia. Taken together, these data suggest that altered mGluR1α and RGS4 mRNA levels may be common in schizophrenia and may represent a disturbed “molecular hub” that has an important effect on multiple components of neuronal communication in the prefrontal cortex.
Increased mGluR1α mRNA levels appear to be specific to the disease process of schizophrenia, or at least not attributable to factors frequently associated with the illness. For example, treatment with antipsychotic medications did not appear to affect mGluR1α mRNA levels, since these levels were similar in schizophrenia subjects who were and were not receiving medications at the time of death. In addition, in contrast to our findings in schizophrenia subjects, mGluR1α mRNA levels were slightly lower in antipsychotic-exposed monkeys, although these differences did not achieve statistical significance. Furthermore, differences in sex; suicide as a cause of death; diagnosis of substance abuse or dependence current at the time of death; the use of antidepressants, benzodiazepines, or sodium valproate at the time of death; or a history of cannabis use did not appear to affect mGluR1α levels in schizophrenia subjects. Interestingly, mGluR1α mRNA levels and mGluR5 mRNA levels appeared to be higher in schizophrenia subjects than in schizoaffective disorder subjects. However, no other markers showed a difference between schizophrenia and schizoaffective disorder subjects, including the cannabinoid 1 receptor (25).
Several lines of evidence suggest that the capacity for mGluR1α activation may be increased in the prefrontal cortex in schizophrenia. First, our findings of elevated mGluR1α mRNA levels parallel a previous report of higher mGluR1α protein levels in the prefrontal cortex in schizophrenia (12), suggesting that higher levels of mGluR1α mRNA expression may lead to translation of more protein, although this hypothesis needs to be directly tested in the same subjects. Second, RGS4 reduces the duration of intracellular signaling from mGluR1α (13). Consistent with evidence that RGS4 also regulates G protein coupling for multiple other receptors (36–39), RGS4 and mGluR1α mRNA levels were not correlated in individual subjects, and differences in RGS4 and mGluR1α mRNA levels between schizophrenia and matched normal comparison subjects were not correlated in subject pairs. However, out of the 42 subject pairs, 27 pairs showed both higher mGluR1α and lower RGS4 mRNA levels in the schizophrenia subject relative to the matched normal comparison subject. Taken together, these data suggest that higher mGluR1α and lower RGS4 mRNA levels are generally found in the same schizophrenia subjects and, if accompanied by changes in levels of the corresponding proteins, may lead to a greater capacity for, and a longer duration of intracellular signaling from, mGluR1α activation in the prefrontal cortex in schizophrenia (Figure 4).
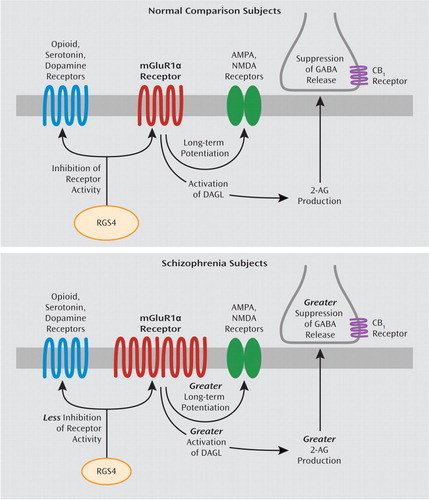
FIGURE 4. Potential Downstream Effects of Alterations in mGluR1α and RGS4 mRNA Levels on Neurotransmitter Systems in Schizophreniaa
a The top image shows that under normal conditions, activation of mGluR1α results in long-term potentiation of NMDA and AMPA receptors (11) and activation of diacylglycerol lipase (DAGL), which leads to synthesis of 2-arachidonoylglycerol (2-AG) (24) and suppression of GABA release from nearby inhibitory axon terminals that contain the cannabinoid 1 receptor (CB1) (6–10). In addition, RGS4 reduces signaling through several different G protein-coupled receptors, including group I mGluRs and opioid, serotonin, and dopamine receptors (13, 36–39). The bottom image shows that in schizophrenia, higher mGluR1α and lower RGS4 mRNA levels suggest the presence of enhanced signaling through mGluR1α. Higher mGluR1α signaling may have diverse effects on multiple components of neural transmission in schizophrenia, including greater long-term potentiation of NMDA and AMPA receptors, enhanced 2-AG synthesis, and greater suppression of GABA release from inhibitory axon terminals that contain the CB1 receptor. In addition, lower RGS4 levels may result in less inhibition of several classes of G protein-coupled receptors. However, additional studies characterizing the cell-type specificity of transcript and protein-level alterations in mGluR1α and RGS4 are needed to clarify the nature of prefrontal cortex circuitry disturbances in schizophrenia.
As previously mentioned, alterations in RGS4 mRNA levels in schizophrenia may have additional effects beyond modifying the intracellular signaling efficacy of mGluR1α. First, RGS4 is a GTPase-activating protein that reduces the duration of activity of several different G protein-coupled receptors in addition to mGluRs, including dopamine, serotonin, and opioid receptors (36–39). Thus, lower RGS4 mRNA levels, if also present at the protein level (16), may enhance the functioning of an array of receptors in schizophrenia (15) (Figure 4). Second, some, although not all (40), case-control and family-based association studies have identified single nucleotide polymorphisms (SNPs) of the RGS4 gene as susceptibility factors for schizophrenia, although different alleles of these SNPs have been associated with schizophrenia in different subject populations (14, 41). Some of the risk haplotypes for RGS4 are also associated with smaller volume of the prefrontal cortex (42) and altered activation of the prefrontal cortex during working memory tasks (43) in schizophrenia subjects. Thus, alterations in RGS4 at the genomic and transcript level may have a broad effect on diverse neurotransmitter systems and on functional properties of the prefrontal cortex in schizophrenia.
Higher mRNA levels for mGluR1α and lower mRNA levels for RGS4, if accompanied by altered levels of the corresponding proteins (12, 16), are consistent with a higher capacity for mGluR1α-mediated neurotransmission in schizophrenia, which may have a diverse and complicated effect on multiple components of neural transmission in the disorder. For example, immunoreactivity for mGluR1α and RGS4 is found in both pyramidal neurons and GABA neurons in the primate prefrontal cortex (44, 45). An important limitation of our study is that a tissue-level analysis of mRNA levels does not allow a determination of a potential cell-type specificity of higher mGluR1α and lower RGS4 mRNA levels in schizophrenia, and thus follow-up studies are needed to determine whether altered levels of mGluR1α and RGS4 are predominantly found in pyramidal neurons or GABA neurons or both. With this caveat in mind, we will conclude by focusing on the potential downstream effect of greater mGluR1α-mediated neurotransmission on the NMDA receptor, endocannabinoid, and GABA systems in schizophrenia. For example, activation of group I mGluRs results in long-term potentiation of NMDA receptor function (11), and NMDA receptor function appears to be impaired in schizophrenia (5). Thus, these lines of evidence invite speculation that in schizophrenia, greater mGluR1α activity may have a partially compensatory effect for NMDA receptor hypofunction. In addition, activation of group I mGluRs has been reported to increase synthesis of 2-arachidonoylglycerol (24) and suppress GABA release (6–10). mGluR1α-mediated activation of 2-arachidonoylglycerol synthesis has been reported to occur through a G protein-coupled mechanism that leads to increased levels of substrate (i.e., diacylglycerol) for diacylglycerol lipase (46). Therefore, while mRNA levels for the synthesizing and metabolizing enzymes for 2-arachidonoylglycerol are not altered in schizophrenia, higher mGluR1α activity may increase the substrate-dependent activity of diacylglycerol lipase, even in the absence of changes in its enzyme level. Thus, higher mGluR1α-mediated activation of 2-arachidonoylglycerol synthesis may further suppress GABA release, which may worsen GABA-ergic deficits in schizophrenia. Higher mGluR1α mRNA levels and lower RGS4 mRNA levels may also affect additional components of neural transmission, including AMPA, dopamine, serotonin, and opioid receptors (11, 36–39) (Figure 4). Taken together, these data suggest that altered mGluR1α and RGS4 mRNA levels may represent a disturbed molecular hub that has an important effect on multiple neurotransmitter systems in schizophrenia. Additional studies characterizing the cell-type specificity of transcript and protein level alterations in mGluR1α and RGS4 may provide greater insight into their downstream effects on prefrontal cortex circuitry, and potentially their relationship to cognitive impairments, in schizophrenia.
1. : What are the functional consequences of neuro-cognitive deficits in schizophrenia? Am J Psychiatry 1996; 153:321–330Link, Google Scholar
2. : Cognitive impairment in schizophrenia is the core of the disorder. Crit Rev Neurobiol 2000; 14:1–21Crossref, Medline, Google Scholar
3. : Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci 2005; 6:312–324Crossref, Medline, Google Scholar
4. : Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol Psychiatry 2008; 13:147–161Crossref, Medline, Google Scholar
5. : Circuit-based framework for understanding neurotransmitter and risk gene interactions in schizophrenia. Trends Neurosci 2008; 31:234–242Crossref, Medline, Google Scholar
6. : Evidence for metabotropic glutamate receptor activation in the induction of depolarization-induced suppression of inhibition in hippocampal CA1. J Neurosci 1998; 18:4870–4882Crossref, Medline, Google Scholar
7. : Metabotropic gluta-mate receptors drive the endocannabinoid system in hippo-campus. J Neurosci 2001; 21:RC188Crossref, Medline, Google Scholar
8. : Cooperative endocannabinoid production by neuronal depolarization and group I metabotropic glutamate receptor activation. Eur J Neurosci 2002; 15:953–961Crossref, Medline, Google Scholar
9. : Multiple mechanisms of endocannabinoid response initiation in hippocampus. J Neurophysiol 2006; 95:67–75Crossref, Medline, Google Scholar
10. : Heterosynaptic LTD of hippocampal GABAergic synapses: a novel role of endocannabinoids in regulating excitability. Neuron 2003; 38:461–472Crossref, Medline, Google Scholar
11. : Metabotropic glutamate receptor-dependent long-term potentiation. Neuropharmacology 2009; 56:735–740Crossref, Medline, Google Scholar
12. : Metabotropic glutamate receptor protein expression in the prefrontal cortex and striatum in schizophrenia. Synapse 2005; 57:123–131Crossref, Medline, Google Scholar
13. : RGS4 inhibits signaling by group I metabotropic glutamate receptors. J Neurosci 1998; 18:905–913Crossref, Medline, Google Scholar
14. : Association and linkage analyses of RGS4 polymorphisms in schizophrenia. Hum Mol Genet 2002; 11:1373–1380Crossref, Medline, Google Scholar
15. : Disease-specific changes in regulator of G-protein signaling 4 (RGS4) expression in schizophrenia. Mol Psychiatry 2001; 6:293–301Crossref, Medline, Google Scholar
16. : Regional alterations in RGS4 protein in schizophrenia. Synapse 2006; 59:472–479Crossref, Medline, Google Scholar
17. : A second endogenous cannabinoid that modulates long-term potentiation. Nature 1997; 388:773–778Crossref, Medline, Google Scholar
18. : Localization of diacylglycerol lipase-alpha around postsynaptic spine suggests close proximity between production site of an endocannabinoid, 2-arachidonoyl-glycerol, and presynaptic cannabinoid CB1 receptor. J Neurosci 2006; 26:4740–4751Crossref, Medline, Google Scholar
19. : Molecular composition of the endocannabinoid system at glutamatergic synapses. J Neurosci 2006; 26:5628–5637Crossref, Medline, Google Scholar
20. : Presynaptically located CB1 cannabinoid receptors regulate GABA release from axon terminals of specific hippocampal interneurons. J Neurosci 1999; 19:4544–4558Crossref, Medline, Google Scholar
21. : The CB1 cannabinoid receptor is the major cannabinoid receptor at excitatory pre-synaptic sites in the hippocampus and cerebellum. J Neurosci 2006; 26:2991–3001Crossref, Medline, Google Scholar
22. : Endocannabinoid signaling as a synaptic circuit breaker in neurological disease. Nat Med 2008; 14:923–930Crossref, Medline, Google Scholar
23. : Brain monoglyceride lipase participating in endocannabinoid inactivation. Proc Natl Acad Sci U S A 2002; 99:10819–10824Crossref, Medline, Google Scholar
24. : Stimulation of endocannabinoid formation in brain slice cultures through activation of group I metabotropic glutamate receptors. Mol Pharmacol 2005; 68:1196–1202Crossref, Medline, Google Scholar
25. : Reduced cortical cannabinoid 1 receptor messenger RNA and protein expression in schizophrenia. Arch Gen Psychiatry 2008; 65:772–784Crossref, Medline, Google Scholar
26. : Regional distribution and effects of postmortal delay on endocannabinoid content of the human brain. Neuroscience 2008; 152:1032–1039Crossref, Medline, Google Scholar
27. : Decreased dendritic spine density on pre-frontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry 2000; 57:65–73Crossref, Medline, Google Scholar
28. : Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res 2005; 33:1–12Crossref, Medline, Google Scholar
29. : Decreased glutamic acid decarboxylase67 messenger RNA expression in a subset of prefrontal cortical gamma-aminobutyric acid neurons in subjects with schizophrenia. Arch Gen Psychiatry 2000; 57:237–245Crossref, Medline, Google Scholar
30. : Conserved regional patterns of GABA-related transcript expression in the neocortex of subjects with schizophrenia. Am J Psychiatry 2008; 165:479–489Link, Google Scholar
31. : Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002; 3:34.1–34.11Crossref, Google Scholar
32. : Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain. J Cell Biol 2003; 163:463–468Crossref, Medline, Google Scholar
33. : The influence of chronic exposure to antipsychotic medications on brain size before and after tissue fixation: a comparison of haloperidol and olanzapine in macaque monkeys. Neuropsychopharmacology 2005; 30:1649–1661Crossref, Medline, Google Scholar
34. : Applied Linear Statistical Models, 4th ed, Vol 1. Boston, McGraw-Hill, 1996Google Scholar
35. : CB1 cannabinoid receptor activity is modulated by the cannabinoid receptor interacting protein CRIP 1a. Mol Pharmacol 2007; 72:1557–1566Crossref, Medline, Google Scholar
36. : GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein alpha subunits. Cell 1996; 86:445–452Crossref, Medline, Google Scholar
37. : Differential effects of regulator of G protein signaling (RGS) proteins on serotonin 5-HT1A, 5-HT2A, and dopamine D2 receptor-mediated signaling and adenylyl cyclase activity. Cell Signal 2004; 16:711–721Crossref, Medline, Google Scholar
38. : RGS4 inhibits Gq-mediated activation of mitogen-activated protein kinase and phosphoinositide synthesis. J Biol Chem 1997; 272:11924–11927Crossref, Medline, Google Scholar
39. : Regulator of G protein signaling 4 confers selectivity to specific G proteins to modulate mu- and delta-opioid receptor signaling. Cell Signal 2009; 21:1218–1228Crossref, Medline, Google Scholar
40. : No significant association of 14 candidate genes with schizophrenia in a large European ancestry sample: implications for psychiatric genetics. Am J Psychiatry 2008; 165:497–506Link, Google Scholar
41. : Evaluation of a susceptibility gene for schizophrenia: genotype based meta-analysis of RGS4 polymorphisms from thirteen independent samples. Biol Psychiatry 2006; 60:152–162Crossref, Medline, Google Scholar
42. : Genetic polymorphisms of the RGS4 and dorsolateral prefrontal cortex morphometry among first episode schizophrenia patients. Mol Psychiatry 2005; 10:213–219Crossref, Medline, Google Scholar
43. : Allelic variation in RGS4 impacts functional and structural connectivity in the human brain. J Neurosci 2007; 27:1584–1593Crossref, Medline, Google Scholar
44. : Distribution of mGluR1alpha and m GluR5 immunolabeling in primate prefrontal cortex. J Comp Neurol 2003; 467:521–535Crossref, Medline, Google Scholar
45. : Mapping the regulator of G protein signaling 4 (RGS4): presynaptic and postsynaptic substrates for neuroregulation in prefrontal cortex. Cereb Cortex 2009; 19:2145–2155Crossref, Medline, Google Scholar
46. : Endocannabinoid-mediated synaptic plasticity in the CNS. Annu Rev Neurosci 2006; 29:37–76Crossref, Medline, Google Scholar



