Neurobiological Correlates of Change in Adaptive Behavior in Autism
Abstract
Objective:
Autism spectrum disorder (ASD) is a lifelong neurodevelopmental condition that is associated with significant difficulties in adaptive behavior and variation in clinical outcomes across the life span. Some individuals with ASD improve, whereas others may not change significantly, or regress. Hence, the development of “personalized medicine” approaches is essential. However, this requires an understanding of the biological processes underpinning differences in clinical outcome, at both the individual and subgroup levels, across the lifespan.
Methods:
The authors conducted a longitudinal follow-up study of 483 individuals (204 with ASD and 279 neurotypical individuals, ages 6–30 years), with assessment time points separated by ∼12–24 months. Data collected included behavioral data (Vineland Adaptive Behavior Scale–II), neuroanatomical data (structural MRI), and genetic data (DNA). Individuals with ASD were grouped into clinically meaningful “increasers,” “no-changers,” and “decreasers” in adaptive behavior. First, the authors compared neuroanatomy between outcome groups. Next, they examined whether deviations from the neurotypical neuroanatomical profile were associated with outcome at the individual level. Finally, they explored the observed neuroanatomical differences’ potential genetic underpinnings.
Results:
Outcome groups differed in neuroanatomical features (cortical volume and thickness, surface area), including in “social brain” regions previously implicated in ASD. Also, deviations of neuroanatomical features from the neurotypical profile predicted outcome at the individual level. Moreover, neuroanatomical differences were associated with genetic processes relevant to neuroanatomical phenotypes (e.g., synaptic development).
Conclusions:
This study demonstrates, for the first time, that variation in clinical (adaptive) outcome is associated with both group- and individual-level variation in anatomy of brain regions enriched for genes relevant to ASD. This may facilitate the move toward better targeted/precision medicine approaches.
Autism spectrum disorder (ASD) is a lifelong neurodevelopmental condition. It is diagnosed behaviorally based on symptoms in the domains of social communication, restricted and repetitive patterns of behaviors and interests, and sensory anomalies (1). These symptoms affect adaptive behavior, that is, the development and application of the abilities required for the attainment of personal independence and social sufficiency (2, 3). They can have severe consequences for individuals and their families. For instance, only 12% of autistic adults are in full-time paid work and only 5% get married (4). Consequently, adaptive behavior is often a key outcome measure of clinical trials in ASD (5–7).
However, across the lifespan, some individuals with ASD improve in terms of adaptive behavior (8), while others may not change significantly (8) or regress (9). The drivers of this variable change remain unclear. This is problematic, especially for the development of effective pharmacological interventions. In particular, the substantial biological and clinical heterogeneity in ASD have contributed to “one-size-fits-all” clinical trials failing to identify treatment effects in this condition. To overcome this challenge and to move toward more targeted (“precision medicine”) interventions, we need to uncover the biological processes that underpin clinical outcome (e.g., change in adaptive behavior, or adaptive outcome), in both subgroups and individuals with ASD.
Notably, given the developmental nature of ASD, the drivers of behavioral change in this condition may also vary across childhood and young adulthood (3). Therefore, to help establish the context of use in which our findings may apply, it is crucial to examine the biological processes underpinning adaptive outcome across age, that is, from childhood to adulthood.
Currently, the best predictor of poor adaptive outcome in ASD is low overall cognitive functioning (IQ) (10, 11). However, a significant proportion of autistic individuals with average or above-average IQ also have poor long-term social outcomes (e.g., 12). Hence, we need to identify additional drivers of adaptive outcome, such as brain function and neuroanatomy. There is preliminary evidence that variation in brain function—that is, resting-state functional connectivity in specific networks—may predict adaptive outcome in ASD (13). That finding constitutes an important first step, but the study had a moderate sample size (N=31), only included male adolescents and young adults without intellectual disability, and focused on brain function at a group level. Moreover, several studies have reported a link between neuroanatomy (e.g., amygdala volume [14], local gray matter volume [15], and white matter integrity [16]) in children with ASD and cross-sectional measures of adaptive behavior. Further, we and others recently linked variability in neuroanatomical features to variation in genes that are mechanistically relevant to these features (17, 18). However, no study to date has examined the neuroanatomical (and associated genetic) correlates of change in adaptive behavior at both the subgroup and individual levels across age.
This is partly because suitable longitudinal deep-phenotyping data within ASD have previously been scarce. Now, the EU-AIMS Longitudinal European Autism Project (LEAP) (19) allows us to examine this question for the first time in one of the largest longitudinal ASD samples worldwide. We included data from 483 individuals (204 with ASD and 279 neurotypical individuals, 6–30 years of age) at two time points separated by ∼12–24 months (T1 and T2), and collected adaptive behavioral data (Vineland Adaptive Behavior Scale–II [VABS-II] [20], at T1 and T2), neuroanatomical data (MRI, at T1), and genetic data (DNA, at T1). See Figure 1 for a study overview.
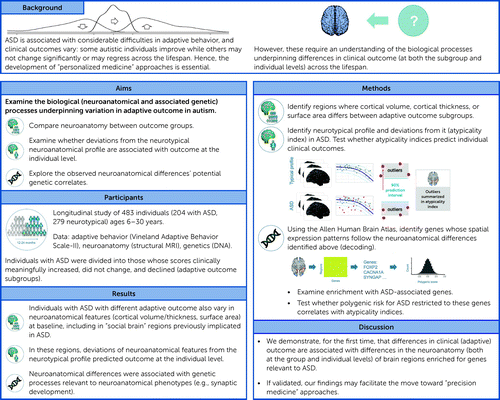
FIGURE 1. Graphical overview of the study
Methods
Participants
Participants were selected from the Longitudinal European Autism Project (LEAP) (described, e.g., in reference 21). The final sample included 204 individuals with ASD and 279 neurotypical individuals. The study was approved by national/local ethics review boards at each study site and carried out to the International Council for Harmonization’s Good Clinical Practice standards. See the online supplement for a full description of inclusion and exclusion criteria, clinical assessments, medication status, and ethical approval boards. Study subjects or their parents or guardians provided informed written or verbal consent after receiving a complete description of the study.
Adaptive Behavior Captured Using the VABS-II
To assess our primary measure (autistic participants’ adaptive behavior), we interviewed their parents/carers using the VABS-II (20), which captures a person’s current level of everyday functioning across multiple domains. We chose this measure because over the past 15 years, the VABS has been widely used as a primary instrument to capture adaptive behavior, including in ASD. We computed Adaptive Behavior Composite (ABC) summary or standard scores (T1, T2, change between visits: VT2−VT1) and used recently published estimates of what constitutes a “minimal clinically important difference” (MCID) (22) to classify autistic individuals into three adaptive outcome subgroups: those whose scores could be said to meaningfully improve (“increasers”; VT2−VT1 ≥ 4), those who showed no meaningful change (“no-changers”; −4 < VT2−VT1 < 4), and those whose scores meaningfully declined (“decreasers”; −4 ≥ VT2−VT1). (See the online supplement and Figure S1 in the supplement.)
MRI Data Acquisition
Using standard 3-T MRI scanners, we acquired high-resolution T1-weighted volumetric structural images with full-head coverage (field of view=27 cm, slice thickness=1.2 mm, in-plane resolution=1.1×1.1 mm2; more detail is provided in Table S1 in the online supplement). Consistent image quality was ensured through both manual and automated quality control procedures.
Cortical Reconstruction Using FreeSurfer
Neuroimaging data were preprocessed using well-validated, fully automated procedures (see the online supplement). From an original sample of 709 individuals (416 with ASD and 293 neurotypical individuals), we retained data from 483 individuals (204 with ASD and 279 neurotypical individuals) (Table 1) and computed vertex-wise measures of three cortical morphometric features: cortical volume, cortical thickness, and surface area.
| Increase Group (N=78) | No-Change Group (N=62) | Decrease Group (N=64) | Within-Group Comparison (ASD) | ASD (N=204) | Neurotypical Group (N=279) | Between-Group Comparison (ASD vs. Neurotypical) (N=483) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Measure | Mean | SD | N | Mean | SD | N | Mean | SD | N | Statistic | df | p | Mean | SD | N | Mean | SD | N | Statistic | df | p |
| ADHD inatt | 3.7 | 2.8 | 67 | 4.9 | 3.6 | 56 | 4.7 | 3.2 | 59 | 2.439 | 2, 179 | 0.090 | 4.37 | 3.2 | |||||||
| ADHD hyper/imp | 1.7 | 2.3 | 67 | 3.6 | 3.1 | 56 | 2.6 | 2.8 | 59 | 6.946 | 2, 179 | 0.001 | 2.6 | 2.8 | |||||||
| ADI total | 34.3 | 12.7 | 77 | 38.0 | 12.3 | 62 | 33.8 | 13.6 | 63 | 2.061 | 2, 199 | 0.090 | 35.3 | 13.0 | |||||||
| ADI social | 16.8 | 6.7 | 77 | 18.2 | 6.1 | 14.5 | 7.0 | 63 | 1.136 | 2, 199 | 0.323 | 17.1 | 6.6 | ||||||||
| ADI comm | 13.6 | 5.6 | 77 | 14.9 | 5.6 | 13.4 | 5.8 | 63 | 1.433 | 2, 199 | 0.241 | 13.9 | 5.7 | ||||||||
| ADI RRB | 3.9 | 2.4 | 77 | 4.9 | 2.6 | 3.9 | 2.7 | 63 | 3.365 | 2, 199 | 0.037 | 4.2 | 2.6 | ||||||||
| Age (years) | 17.6 | 4.8 | 14.6 | 4.4 | 16.9 | 6.3 | 5.837 | 2, 201 | 0.003 | 16.5 | 5.3 | 17.3 | 5.9 | 2.843 | 1, 481 | 0.092 | |||||
| CSS total | 4.8 | 2.5 | 76 | 5.8 | 2.9 | 61 | 5.4 | 2.9 | 2.091 | 2, 198 | 0.126 | 5.3 | 2.8 | ||||||||
| CSS SA | 5.5 | 2.5 | 76 | 6.4 | 2.6 | 61 | 6.0 | 2.8 | 2.161 | 2, 198 | 0.118 | 5.9 | 2.7 | ||||||||
| CSS RRB | 4.4 | 2.8 | 76 | 4.9 | 2.8 | 61 | 5.1 | 2.9 | 0.871 | 2, 198 | 0.420 | 4.8 | 2.8 | ||||||||
| Full-scale IQb | 104.6 | 17.1 | 102.4 | 21.9 | 96.9 | 19.7 | 2.900 | 2, 201 | 0.057 | 101.5 | 19.6 | 104.8 | 18.2 | 3.619 | 1, 481 | 0.058 | |||||
| Sex | |||||||||||||||||||||
| Female | 21 | 14 | 29 | 8.720 | 2 | 0.013 | 64 | 101 | 115.375 | 1 | 0.019 | ||||||||||
| Male | 57 | 48 | 35 | 140 | 178 | ||||||||||||||||
| VABS | |||||||||||||||||||||
| T1 comm | 74.0 | 12.7 | 75.3 | 13.6 | 81.3 | 17.4 | 4.886 | 2, 201 | 0.008 | 76.7 | 14.8 | ||||||||||
| T1 daily living | 71.6 | 12.0 | 74.5 | 16.2 | 77.5 | 18.2 | 2.593 | 2, 201 | 0.077 | 74.3 | 15.6 | ||||||||||
| T1 social | 70.6 | 15.2 | 70.1 | 14.3 | 73.6 | 15.0 | 1.059 | 2, 201 | 0.349 | 71.4 | 14.9 | ||||||||||
| T1 ABC | 69.6 | 10.4 | 71.4 | 12.2 | 75.4 | 14.5 | 4.008 | 2, 201 | 0.020 | 72.0 | 12.5 | ||||||||||
| VT2−VT1 comm | 9.0 | 12.1 | −2.7 | 6.6 | −14.5 | 12.7 | 80.979 | 2, 201 | ≤0.001 | −2.0 | 14.7 | ||||||||||
| VT2−VT1 daily living | 8.6 | 9.1 | 0.4 | 7.6 | −9.5 | 8.6 | 79.196 | 2, 201 | ≤0.001 | 0.45 | 11.4 | ||||||||||
| VT2−VT1 social | 12.4 | 9.9 | 2.6 | 7.2 | −7.2 | 9.8 | 80.636 | 2, 201 | ≤0.001 | 3.2 | 12.2 | ||||||||||
| VT2−VT1 ABC | 9.8 | 5.3 | 0.2 | 2.0 | −10.5 | 7.6 | 241.474 | 2, 201 | ≤0.001 | 0.5 | 10.1 | ||||||||||
| T2 comm | 82.9 | 14.3 | 72.6 | 12.4 | 66.8 | 20.4 | 18.789 | 2, 201 | ≤0.001 | 74.7 | 17.3 | ||||||||||
| T2 daily living | 80.2 | 12.7 | 74.9 | 16.7 | 68.0 | 15.8 | 11.679 | 2, 201 | ≤0.001 | 74.8 | 15.8 | ||||||||||
| T2 social | 82.9 | 13.5 | 72.7 | 14.3 | 66.4 | 19.0 | 20.285 | 2, 201 | ≤0.001 | 74.6 | 17.1 | ||||||||||
| T2 ABC | 79.4 | 10.5 | 71.5 | 12.5 | 64.9 | 17.4 | 20.199 | 2, 201 | ≤0.001 | 72.5 | 14.8 | ||||||||||
| Total cortical volume (cm3) | 1,207.8 | 122.3 | 1,201.9 | 88.0 | 1,159.9 | 143.3 | 3.143 | 2, 201 | 0.045 | 1,191.0 | 124.5 | 1,189.8 | 126.7 | 0.011 | 1, 481 | 0.917 | |||||
| Mean cortical thickness (mm) | 2.7 | 0.1 | 2.7 | 0.1 | 2.7 | 0.1 | 2.153 | 2, 201 | 0.119 | 2.7 | 0.1 | 2.7 | 0.1 | 4.621 | 1, 481 | 0.032 | |||||
| Total surface area (cm2) | 2,310.6 | 223.0 | 2,307.2 | 184.5 | 2,257.0 | 274.2 | 1.132 | 2, 201 | 0.325 | 2,292.8 | 230.2 | 2,285.3 | 228.5 | 0.124 | 1, 481 | 0.725 | |||||
| Time (months) | 19.6 | 2.3 | 77 | 19.6 | 3.2 | 59 | 19.5 | 3.8 | 63 | 0.028 | 2, 201 | 0.972 | 19.6 | 3.1 | 199 | 19.1 | 3.7 | 212 | 2.188 | 1, 409 | 0.140 |
TABLE 1. Demographic characteristics and total brain measuresa
Neuroimaging Analyses
First, we tested for neuroanatomical differences between adaptive outcome groups at T1. We included adaptive outcome subgroup, sex, and site as fixed-effect factors, and linear (cortical volume, surface area, and cortical thickness) and quadratic (cortical volume and cortical thickness) age (23, 24), IQ, total brain measures (total cortical volume, total surface area, and mean cortical thickness), and T1 ABC scores (VT1) as continuous covariates. We examined between-group differences correcting for multiple comparisons across the whole brain using random field theory–based cluster correction for nonisotropic images with a cluster-defining and cluster p value significance threshold of 0.05 (two-tailed) (25). Given that cortical volume, surface area, and cortical thickness are thought to have different underpinning neurobiological mechanisms (26, 27), we treated them as separate analyses and did not correct for multiple comparisons across these three features.
We conducted supplementary analyses to examine and preclude the influence of several potential confounders. Specifically, we explored the effects of no covariation for total brain, the effect of different corrections for site, the effect of covariation for medication, and the effect of no covariation for IQ.
Also, we extended our primary analyses, which followed a categorical approach based on the potentially clinically meaningful MCID-derived cutoff. Specifically, we conducted supplementary analyses using a continuous approach to test whether there was an association between adaptive outcome and neuroanatomy independent of the MCID cutoff of four units. We also examined the effect of subgroup on neuroanatomy directly across all three groups (rather than using a pairwise approach).
Moreover, our primary analyses harnessed the full age range in our sample. We recognize, however, that given the developmental nature of ASD, the association between neuroanatomy and adaptive outcome may vary across ages and follow-up durations. Therefore, we conducted supplementary analyses controlling for follow-up duration and its interaction with age. To preclude confounding through between-outcome-group differences in age, we further repeated our analyses in a subsample matched for age, sex, and IQ. Additionally, to disentangle the neuroanatomical correlates of adaptive outcome across age, as well as to make our findings more easily comparable to previous research restricted to individual age groups, we conducted supplementary analyses of between-outcome-group differences separately within children/adolescents and adults (we grouped children and adolescents together to achieve balanced numbers of increasers, no-changers, and decreasers across age groups).
Also, our primary goal was to examine adaptive behavior and we relied on a single instrument (VABS-II). Nonetheless, we conducted supplementary analyses to further corroborate an association between neuroanatomy and change in behavior related or relevant to adaptive behavior (such as social communication processing). Specifically, we stratified individuals into outcome groups based on alternative measures of social communication processing in ASD, including total Autism Diagnostic Observation Schedule (ADOS) (28) scores, ADOS social affect domain scores, and Social Responsiveness Scale (SRS) (29) scores. We then compared neuroanatomy between these outcome groups at T1 (see the online supplement). We also examined neuroanatomical differences between all three ASD outcome groups combined and the neurotypical group.
Second, we extended our analyses from the subgroup level to the individual level. We examined whether, in the regions identified above, an individual’s deviation from the neurotypical neurodevelopmental trajectory at T1 (atypicality index) predicted that individual’s subsequent adaptive outcome. We computed atypicality indices for each subject and set of regions and tested whether they predict change in adaptive behavior in ASD, controlling for age, sex, IQ, site, and T1 ABC scores (VT1); we also recomputed our analyses using bootstrapping to increase the accuracy of our estimates (see the online supplement). While these measures are based on regions previously identified, circularity is avoided by examining autistic individuals’ deviation from the neurotypical mean rather than from the mean of other outcome groups.
Genetic Analyses
Next, we investigated how our neuroanatomical findings relate to genomic mechanisms.
Our first analysis (the decoding analysis) was based on the Allen Human Brain Atlas (30). The Allen atlas is the most comprehensive gene expression atlas currently available, but it is based on adult donors only, and its spatial coverage and resolution are much lower than those of neuroimaging data. Consequently, future studies should repeat our decoding analysis using age-specific high-resolution gene expression data sets once they become available. Here, we used the Allen Human Brain Atlas to identify genes that are expressed throughout the brain in spatial patterns similar to the observed neuroanatomical subgroup differences.
After identification of these genes, we investigated 1) their implication in ASD (gene enrichment analysis), 2) their functional roles and biological relevance (protein-protein interaction analysis), and 3) whether variation in these genes correlated with neuroanatomical variability in our participants (see the online supplement).
We tested whether the identified genes may be implicated in ASD by examining whether they were enriched for different classes of genes previously associated with this condition at the genetic/transcriptomic level (see the online supplement). First, we conducted our enrichment analyses using a background list of 20,787 genes, which are all genes considered in Neurosynth (https://neurosynth.org/). Second, to avoid biasing our findings toward genes expressed in brain, we limited our background list to 16,906 genes, based on real estimates of genes expressed in cortical tissue (31) (see Figure S33 in the online supplement). Our enrichment analyses yielded enrichment odds ratios, hypergeometric p values, and false discovery rate (FDR) q-values. Only those tests with pFDR<0.05 were interpreted further.
Next, to explore the functional roles and biological relevance of the identified genes, we conducted protein-protein interaction analyses. In simple terms, these analyses assessed whether the products of the identified genes are thought to interact physically to support biological processes, and if so, which processes they are. Specifically, we tested enrichment for Gene Ontology (GO) biological processes. We then selected enrichments that survived statistical thresholding (pFDR<0.05) and categorized them based on terms relevant to our cortical phenotypes (e.g., autophagy, cell cycle, cell death, growth, neurogenesis, and synapse).
Last, we tested whether variation in the identified genes correlated with neuroanatomical variability in our participants. To do that, we generated polygenic scores (PGSs) for each person and set of neuroanatomical regions (those identified above, i.e., neuroanatomical subgroup differences). In brief, each PGS summarized an individual’s amount of ASD-associated genetic variation within the genes thought to be expressed in that set of regions. The increasers-versus-no-changers neuroanatomical contrast for surface area yielded a very small number of genes (11) in the decoding step; hence, while these results are included for completeness, they should be interpreted with caution (see the online supplement). We then tested the association between PGSs and atypicality indices for each set of regions using Pearson correlation analyses, controlling for age, IQ, sex, site, and the first five genetic principal components (pFDR<0.05). We also repeated our analyses using bootstrapping (4,000 iterations).
Results
Demographic Characteristics
ASD subgroups did not differ significantly in baseline full-scale IQ, ADOS (32), and Autism Diagnostic Interview (ADI) (33) measures (except the ADI restricted and repetitive behavior subscale), inattention (1), daily living, and socialization domain scores (20) at baseline, total surface area, mean cortical thickness, and time between visits. Subgroups differed significantly in T1 adaptive behavior communication domain and ABC scores, all (composite and domain) VT2−VT1 and T2 adaptive behavior scores, age, sex, impulsivity (parent measure), and total cortical volume (Tables 1 and 2) (for medication, see Table S4 in the online supplement). Also, neurotypical and autistic individuals did not differ significantly in age, IQ, total cortical volume, total surface area, and the time between visits, but differed in sex distribution, and mean cortical thickness was larger in the ASD group than in the neurotypical group (Table 1).
| ID | H | Region | T (x, y, z) | T | Vertices | Cluster Probability | Brodmann Area | ||
|---|---|---|---|---|---|---|---|---|---|
| Cortical volume | |||||||||
| Increasers > decreasers | |||||||||
| 1 | R | Precuneus cortex | −10 | −46 | 45 | 2.97 | 3,599 | 1.97 e−04 | 31 |
| Superior parietal cortex | |||||||||
| Isthmus cingulate cortex | |||||||||
| 2 | R | Precuneus cortex | 18 | −34 | 49 | 3.13 | 3,019 | 6.54 e−04 | 5, 7 |
| Superior parietal cortex | |||||||||
| Paracentral lobule | |||||||||
| 3 | R | Superior parietal cortex | 47 | −27 | 37 | 3.47 | 3,343 | 3.00 e−03 | 5, 40 |
| Supramarginal gyrus | |||||||||
| Inferior parietal cortex | |||||||||
| 4 | L | Entorhinal cortex | −32 | −3 | −20 | 3.31 | 433 | 7.12 e−03 | 28 |
| Increasers < decreasers | |||||||||
| 5 | R | Superior frontal gyrus | 8 | 34 | 41 | −1.66 | 2,063 | 1.51 e−03 | 8 |
| 6 | R | Superior temporal gyrus | 59 | −29 | 3 | −1.66 | 2,795 | 5.94 e−03 | 22 |
| Banks of superior temporal sulcus | |||||||||
| 7 | L | Lateral occipital cortex | −41 | −82 | 4 | −1.66 | 2,436 | 7.43 e−03 | 18, 19 |
| Inferior parietal cortex | |||||||||
| 8 | R | Lateral occipital cortex | 30 | −78 | 8 | −1.66 | 1,990 | 2.13 e−02 | 19 |
| Decreasers > no-changers | |||||||||
| 1 | R | Posterior cingulate cortex | 5 | −9 | 28 | −1.66 | 382 | 9.97 e−06 | 23, 31 |
| 2 | L | Superior frontal gyrus | −20 | 57 | 2 | −1.66 | 2,386 | 7.59 e−04 | 10 |
| Rostral middle frontal gyrus | |||||||||
| 3 | R | Superior frontal gyrus | 8 | 43 | 21 | −1.66 | 1,460 | 3.35 e−02 | 9, 11, 12 |
| Cortical thickness | |||||||||
| Increasers > decreasers | |||||||||
| 1 | L | Insula | −36 | −2 | −11 | 3.90 | 1,981 | 9.80 e−06 | 28, 34, 38, 22 |
| Entorhinal cortex | |||||||||
| Inferior temporal gyrus | |||||||||
| Temporal pole | |||||||||
| Superior temporal gyrus | |||||||||
| 2 | R | Precuneus | 25 | −7 | −27 | 3.87 | 428 | 1.03 e−05 | 7 |
| Increasers < decreasers | |||||||||
| 3 | L | Lateral occipital cortex | −21 | −86 | −3 | −1.66 | 2,740 | 7.38 e−04 | 18, 19, 37 |
| Fusiform gyrus | |||||||||
| Surface area | |||||||||
| Increasers > decreasers | |||||||||
| 1 | R | Postcentral gyrus | 59 | −15 | 24 | 3.33 | 8,291 | 3.42 e−05 | 10, 13, 40, 44 |
| Supramarginal gyrus | |||||||||
| Precentral gyrus | |||||||||
| Pars opercularis | |||||||||
| Insula | |||||||||
| Rostral middle frontal gyrus | |||||||||
| 2 | R | Precuneus cortex | 14 | −30 | 46 | 4.11 | 4,797 | 3.20 e−03 | 5, 7 |
| Paracentral lobule | |||||||||
| Superior parietal cortex | |||||||||
| 3 | L | Inferior temporal gyrus | −22 | −26 | −19 | 2.80 | 3,092 | 3.21 e−02 | 20, 36, 34, 37 |
| Fusiform gyrus | |||||||||
| Parahippocampal gyrus | |||||||||
| 4 | L | Supramarginal gyrus | −43 | −29 | 33 | 2.78 | 4,929 | 4.07 e−02 | 1, 2, 3, 40 |
| Postcentral gyrus | |||||||||
| Increasers < decreasers | |||||||||
| 5 | R | Lingual gyrus | 19 | −34 | −5 | −1.66 | 1,743 | 2.14 e−02 | 19, 30, 36 |
| Isthmus cingulate cortex | |||||||||
| Increasers > no-changers | |||||||||
| 1 | R | Lingual gyrus | 17 | −73 | 13 | 3.98 | 3,718 | 9.98 e−06 | 17, 19, 30, 34, 36 |
| Isthmus cingulate cortex | |||||||||
| Pericalcarine cortex | |||||||||
| Parahippocampal gyrus | |||||||||
| 2 | L | Lingual gyrus | −20 | −51 | 6 | 3.50 | 7,332 | 6.38 e−04 | 17, 19, 30, 36 |
| Lateral occipital cortex | |||||||||
| Pericalcarine cortex | |||||||||
| Cuneus cortex | |||||||||
| Isthmus cingulate cortex | |||||||||
| Decreasers < no-changers | |||||||||
| 1 | L | Superior parietal cortex | −34 | −40 | 39 | 3.57 | 5,938 | 6.31 e−03 | 1, 2, 3, 40 |
| Postcentral gyrus | |||||||||
| 2 | L | Precuneus cortex | −15 | −46 | 36 | 3.08 | 4,403 | 1.04 e−02 | 19, 30, 31, 36 |
| Lingual gyrus | |||||||||
| Isthmus cingulate cortex | |||||||||
| Pericalcarine cortex | |||||||||
TABLE 2. Regions displaying significant neuroanatomical baseline differences between those individuals whose adaptive behavioral scores increased, did not change, and decreaseda
Neuroanatomical Differences Between Adaptive Outcome Groups
At baseline, we observed widespread neuroanatomical differences between increasers and decreasers in cortical volume, cortical thickness, and surface area (Figure 2A–C). Increasers and no-changers differed in cortical volume and surface area (Figure 2E,D); and decreasers and no-changers differed in surface area and cortical volume (Figure 2F,G). These results are also summarized in Table 2 and in the online supplement. Main results were corrected for multiple comparisons across the whole brain using random field theory–based cluster correction for nonisotropic images with a cluster-defining and cluster p value significance threshold of <0.05 (two-tailed). (For additional statistical thresholds, see Figures S12–S14 in the online supplement. For uncorrected T-values, see Figures S2 and S3 in the online supplement. For overall model effect sizes, see Figure S4 in the online supplement. For effect sizes of each model term in each between-group contrast or feature, see Figures S5–S11 in the online supplement).
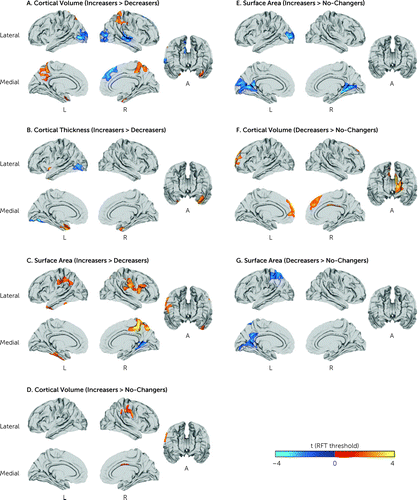
FIGURE 2. Neuroanatomical baseline differences between adaptive outcome groupsa
a Panels A–C show differences between increasers and decreasers in, respectively, cortical volume, cortical thickness, and surface area. Panels D and E show differences between increasers and no-changers in, respectively, cortical volume and surface area. Panels F and G show differences between decreasers and no-changers in, respectively, cortical volume and surface area. T-values are random field theory–corrected and are indicated by color bars. A=anterior view; L=left; R=right; RFT=random field theory.
We conducted extensive supplementary analyses to test the robustness of our findings considering several potential confounding factors. These included total brain measures (Figures S15–S17 in the online supplement); site, using the ComBat batch effect harmonization method (34) or random-effect modeling (see Figures S18–S20 in the online supplement); medication (see Figures S21–S23 in the online supplement); no correction for IQ (see Figure S24 in the online supplement); examining the association between neuroanatomy and adaptive outcome continuously, that is, irrespective of the MCID-based cutoff (see Figure S25 in the online supplement); and focusing on the main effect of outcome group across all groups rather than computing pairwise comparisons (see Figure S26 in the online supplement). Taken together, our findings remained highly consistent when accounting for these factors.
Further, given the developmental nature of ASD, we also explored the association between neuroanatomy and adaptive outcome in view of participants’ age and the timing of visits. Our analyses showed that our results were not confounded by age, follow-up duration, or their interaction (see Figure S27 in the online supplement). Similarly, they did not arise from age differences between outcome groups: our findings remained robust in a subsample matched for age, sex, and IQ (e.g., there were similar differences in frontal cortical volume (increasers < decreasers; decreasers > no-changers); temporal (increasers < no-changers), parietal (decreasers < no-changers), and occipital (increasers > decreasers) surface area; and temporal cortical thickness (increasers > decreasers) (see Figure S28 in the online supplement). Further, to investigate whether variation in age contributes to distinct associations between neuroanatomy and adaptive outcome, we repeated our analyses within children/adolescents and adults. These analyses yielded results similar to those observed across age groups, although between-group differences were greater in younger individuals than in adults (see Figure S29 in the online supplement). For instance, for the increasers-versus-decreasers contrast, there were overlapping differences in frontal, occipital (increasers < decreasers), and parietal (increasers > decreasers) cortical volume; parieto-occipital and temporal (increasers > decreasers) surface area; and temporal cortical thickness (increasers > decreasers) in the children and similarities in fronto-parietal surface area (increasers > decreasers) and temporal cortical thickness (increasers > decreasers). Combined, these findings suggest that in ASD, variation in neuroanatomy and adaptive outcome are associated across the lifespan, and perhaps especially in children/adolescents.
Moreover, we examined whether our findings of an association between neuroanatomy and adaptive outcome (based on the VABS-II) extended to other measures capturing social communication symptoms. We found that outcome groups based on alternative measures (ADOS total, ADOS social affect, SRS) also differed in neuroanatomy at T1, but the spatial patterns displayed low to moderate correlations with those derived from VABS-II-based subgroups (see Figure S30 in the online supplement). Last, for the sake of completeness, we identified neuroanatomical differences between the neurotypical group and all ASD outcome groups combined (see Figure S31 in the online supplement).
Neuroanatomical Atypicality and Individual Change in Adaptive Behavior
In the regions identified above, an individual’s deviation from the neurotypical developmental trajectory predicted their adaptive outcome. For increasers versus decreasers, a greater cortical volume atypicality index predicted a decline in adaptive behavior (p=0.008). In contrast, a more positive surface area atypicality index (increasers vs. decreasers), cortical volume atypicality index (increasers vs. no-changers) and surface area atypicality index (decreasers vs. no-changers) predicted an improvement in adaptive behavior (p≤0.001, p=0.016, and p=0.003, respectively). Importantly, the decreasers displayed greater absolute deviations from the neurotypical group than did the increasers. Hence, greater (absolute) neuroanatomical deviation predicted a worsening in adaptive behavior. Adding atypicality indices as predictors increased the amount of explained variance, even while adjusting for the total number of predictors in the model, from 8.1% (adjusted R2 of the model without atypicality indices) to up to 14.4% (adjusted R2 of the model including atypicality indices). Taken together, our findings suggest that an individual’s adaptive outcome may be predicted more accurately by considering not only demographic and behavioral features (age, IQ, etc.), but also neuroanatomy (see Table S2 in the online supplement; for results derived using bootstrapping, see Table S3 in the online supplement).
Genetic Correlates of Neuroanatomical Subgroup Differences
We also examined the genetic associates of the observed neuroanatomical differences between the adaptive outcome subgroups.
Several of the regions that differed between subgroups displayed an enrichment for genes implicated in ASD. Regions differing between increasers and decreasers in cortical volume were enriched for gene sets previously reported to be transcriptionally downregulated in ASD (35) (odds ratio=3.2, pFDR<0.01). Regions differing between increasers and decreasers in surface area were enriched for genes downregulated in ASD (35) (odds ratio=3.07), coexpression modules downregulated in ASD (36) (odds ratio=2.26), and many genes differentially expressed in bipolar disorder (odds ratio=1.62) and schizophrenia (odds ratio=1.57) (35). In contrast, regions differing between increasers and no-changers in cortical volume were enriched for coexpression modules upregulated in ASD (36) (Figure 3A) (for effect sizes, see Figure S32 in the online supplement). The remaining imaging contrasts showed no significant enrichments. These results remained largely unchanged when using a more restrictive background total of 16,906 genes (genes expressed in cortical tissue) (31) (see Figure S33 in the online supplement).
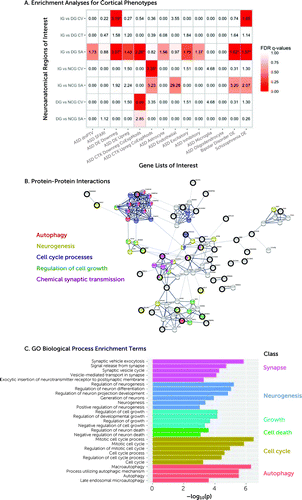
FIGURE 3. Genetic correlates of neuroanatomical baseline differences between adaptive outcome groupsa
a Panel A shows the enrichment analyses for cortical phenotypes (rows) by ASD-associated gene lists (columns). Tile colors indicate false discovery rate (FDR) q-values. Tile labels indicate enrichment odds ratios, and significant values are marked with an asterisk. Panel B shows protein-protein interactions between genes overlapping on the increasers-versus-decreasers cortical volume or surface area lists and the ASD DE Downreg list. Seed genes are denoted with thick black outlines. Other genes are highly connected interactors with the seed gene. Edge thickness indicates confidence in the interaction (thicker=higher confidence). Nodes are colored to indicate genes that are members of important Gene Ontology (GO) biological process terms, such as autophagy, neurogenesis, cell cycle processes, regulation of cell growth, and chemical synaptic transmission. Panel C shows GO biological process enrichment terms from an extended protein-protein interaction network around genes that are relevant to increasers-versus-decreasers cortical volume and surface area and ASD DE Downreg lists. CT=cortical thickness; CV=cortical volume; DG=decrease group; IG=increase group; NCG=no-change group; SA=surface area.
An examination of the functional roles and biological relevance of the genes identified above revealed that the genes associated with the increasers-versus-decreasers contrast and that are downregulated in ASD interact at the protein level (p=1.06×10−6). Specifically, an extended protein-protein interaction network analysis showed that these genes or their products are involved in biological processes that are relevant to the cortical phenotypes of cortical volume and surface area, including neurogenesis, cell division, regulation of cell growth, and mitotic cell cycle processes (Figure 3B,C).
Further, we examined whether variation in these genes correlated with neuroanatomical variability among our participants. We found that in regions where increasers and decreasers differed in cortical thickness, atypicality indices correlated significantly and positively with PGSs (r=0.271, pFDR=0.003, N=142, representing a small to medium effect size; bootstrap: bias=0.000, SE=0.080, 95% CI=0.106, 0.417). Thus, more atypical cortical thickness was accompanied by greater polygenic variation associated with ASD (restricting our analyses to the genes expressed here). We observed no correlations between the atypicality indices and PGSs for other contrasts or features.
Discussion
We examined, for the first time, the neuroanatomical correlates of change in adaptive behavior in ASD over a period of ∼1–2 years at both the group and the individual level. We demonstrate that ASD subgroups with different adaptive outcomes at follow-up were distinguished by widespread neuroanatomical differences at baseline. Moreover, in ASD, at an individual level, absolute deviation from the neurotypical neuroanatomical developmental profile predicted subsequent adaptive outcome. In regions that differed neuroanatomically between ASD subgroups, we discovered 1) enrichment for genes known to be transcriptionally downregulated in cortical tissue in ASD, and 2) an association between deviations from the typical developmental trajectory and polygenic variation (in the genes expressed here) associated with ASD. Notably, in predicting adaptive outcome, neuroanatomical profiles carried weights similar to IQ. Also, subgroups differed neurobiologically but not in IQ. Hence, analyzing phenotypic features other than IQ (i.e., brain anatomy) may help us better understand putative mechanisms underpinning variation in adaptive outcome. Taken together, our study findings are among the first to characterize neuroanatomical (and associated genetic) anomalies associated with adaptive outcome in ASD.
Before discussing our findings and their implications, we will highlight several methodological considerations.
Our study sample covered a broad age range (6–30 years). Taking such a truly dimensional approach without applying arbitrary age barriers allowed us to examine real-life heterogeneity across the autism spectrum. Nonetheless, we recognize that the processes underpinning adaptive behavioral change may vary across developmental stages and follow-up durations. For instance, a 6-year-old may be expected to develop differently over 2 years compared to a 30-year-old. Therefore, we also repeated our analyses covarying for age, follow-up duration, and their interaction. To further preclude confounding through age, we repeated our analyses in a subset of participants matched for age. Last, to disentangle the processes underpinning adaptive outcome across age, and also to make our findings more relevant to previous studies examining individual age groups (e.g., children or adults), we repeated our analyses separately in children/adolescents and adults. Taken together, these analyses revealed associations between neuroanatomy and adaptive outcome in ASD across the lifespan, and perhaps especially in children/adolescents.
Further, our analyses were centered on the VABS-II, a single instrument to assess adaptive behavior. This decision was based on the fact that the VABS-II remains one of the very few measures in ASD with an evidence-based MCID estimate, that is, for which there exists an empirical measure of what constitutes a clinically meaningful change over time (22). Further, the VABS-II measures communication/socialization skill and therefore captures ASD core deficits. This utility of the VABS-II is reflected in its use as a primary endpoint in registrational phase 3 trials in ASD (e.g., NCT03504917). Also, the focus of this work on adaptive behavior follows the prespecified analytic plan of the EU-AIMS project (described in reference 9). Nonetheless, we extended our analyses to also examine the association between neuroanatomy and change in behavior based on alternative measures of social communication symptoms, including the ADOS total and social affect score, and the SRS. Combined, these analyses revealed an association between neuroanatomy and behavioral outcome in ASD regardless of the type of communication/socialization measure. However, and unsurprisingly, neuroanatomical spatial patterns varied across symptom measures.
We were not yet able to replicate our findings by, for example, using other publicly available ASD data sets. This is because other samples do not possess the same breadth of phenotyping, sample heterogeneity, and longitudinal approach from childhood to adulthood as LEAP does (21). We will address this limitation once suitable data sets become available. Also, we have not yet validated our findings and explored their potential in clinical settings. We plan to address this in future work. First, we aim to validate our findings, especially at the case level, using multivariate pattern analysis and machine learning approaches. Second, we will test the utility of our neuroanatomical findings as stratification markers to enrich trials, for example, for individuals likely to have relatively poor outcomes. Third, we will investigate the contexts of use (e.g., in children/adolescents vs. adults) in which our findings may best inform clinical trials.
In view of these considerations, we first identified spatial patterns of neuroanatomical variability associated with change in adaptive behavior at the group level. Existing studies have primarily examined the relationship between adaptive behavior and neuroanatomy cross-sectionally and within well-defined developmental stages (14, 15). Our work extends these analyses by using an accelerated longitudinal study design in a larger sample with a broader age range (including adults). We established that subgroups differed significantly at T1 across several morphometric features (cortical volume, surface area, and cortical thickness), including in frontal, temporal, parietal, and occipital areas. The anatomical differences included regions previously implicated in ASD, for example, the prefrontal cortex, temporal lobe, and precuneus (37). They also comprised regions linked to social communication processing, attention, cognitive control, and motor learning in ASD, such as the parietal cortex, supramarginal gyrus (38, 39), insula (40, 41), lateral occipital cortex (42), and lingual gyrus (43). Also, these results were robust and survived correction for multiple (potentially confounding) methodological, brain, clinical, behavioral, and demographic factors, such as site effects, total brain measures, medication, IQ, age, follow-up duration and its interaction with age, or subgroup differences in age, sex, and IQ. Given our whole-brain approach, correction for multiple comparisons, and exploration of potential confounders, a clustering of differences in social brain regions adds plausibility to the suggestion that they play a key modulatory role in adaptive outcome (including social communication difficulties). Nonetheless, the extension of spatial patterns into regions associated with other cognitive/behavioral functions implicated in ASD (e.g., attention [41]) suggests that mechanisms beyond social communication processing may also contribute to changes in adaptive behavior.
Taken together, our findings indicate an association between adaptive behavior and brain anatomical differences—but these are likely widespread (i.e., not restricted to one particular region). This association was particularly noticeable in children/adolescents, where supplementary analyses revealed greater between-group differences compared to the analyses in adults. These findings are unsurprising given strong previous evidence that the young brain is highly dynamic and plastic but becomes increasingly mature with age (44). Notably, we also identified widespread neuroanatomical differences between outcome groups derived using alternative communication/socialization measures (ADOS total, ADOS social affect domain, and SRS), further corroborating the link between neuroanatomy and variation in subsequent behavioral change. However, correlations between our VABS-II-based and these supplementary findings were low to moderate. This is unsurprising given the difference in the included participants and the fact that the VABS-II, while capturing the social communication domain, is an independent instrument measuring different and/or additional behavioral features compared to the ADOS/SRS (e.g., daily living skills). Notably, this inconsistency between findings derived through the most commonly used measures of social-communication difficulties in ASD further highlights the significant heterogeneity in ASD and the challenges inherent to characterizing the autistic brain. Combined, our findings suggest that at the subgroup level, change in adaptive behavior, especially in early life, but also in adulthood, is underpinned by regional variation in neuroanatomy at baseline. However, to better understand the developmental origins of adaptive outcome, it is important to examine how change in adaptive behavior relates to different morphometric features at the individual level, not just the group level.
Next, we therefore examined whether atypical neuroanatomy at T1 predicted subsequent adaptive outcome in ASD at the individual level. Previous studies have linked neuroanatomical variation to ASD symptoms, including adaptive behavior (45, 46). We built on this work by demonstrating that where subgroups differed neuroanatomically at baseline, an individual’s deviation from the neurotypical profile predicted their subsequent adaptive outcome. Specifically, more abnormal frontal, temporal, parietal, and occipital cortical volume and frontal, temporal, and parietal surface area predicted a decline in adaptive behavior. Adding neuroanatomical predictors to our model (beyond age, IQ, etc.) increased the amount of explained variance in adaptive outcome.
Nonetheless, the implications of our findings remain to be explored. Atypicality indices provide a means to gauge “accumulated” neuroanatomical atypicality (similar to PGSs in genetics). However, as summary indices, they do not differentiate between positive and negative deviations from the neurotypical range or between individual regions. Similarly, they do not allow inferences about the biological mechanisms contributing to these atypicalities. For instance, it is unclear whether deviations above and below the estimated neurotypical range are driven by similar or different biological mechanisms. Also, our analyses do not allow inferences about whether or how the neuroanatomical atypicality quantified by atypicality indices contributes to subsequent clinical outcome. For example, it is unclear whether or how neuroanatomical atypicality in particular regions increases “susceptibility” to a specific adaptive outcome, and whether this “susceptibility” varies depending on which cortical feature is affected. Last, the implications of atypicality index–based findings may vary depending on the sample used to establish the neurotypical developmental trajectory. For instance, a broad age range (as used in our sample), may increase the generalizability but also diminish the temporal specificity and sensitivity of the derived atypicality indices. Taken together, our findings suggest that specific neurodevelopmental processes modulate change in adaptive behavior in ASD, but that it is the amount of (widespread) deviation from neurotypical neuroanatomy that predicts (poor) adaptive outcome—and not abnormalities in one specific brain region per se. Additional studies are needed to determine how neuroanatomical variation affects outcomes in other ASD symptom domains.
The mechanisms underpinning our group- and individual-level neuroanatomical findings are unclear. Previous studies suggest that cortical volume, surface area, and cortical thickness are regulated through different developmental processes (see, e.g., 26, 27), which in turn are modulated through a complex interplay of genes (see, e.g., 47–49) and environmental factors. Accordingly, studies in rodents (50) and humans (17) have linked neuroanatomical variability to genetic variation. To build on this, here we examined the genetic correlates of outcome-related neuroanatomical variability. We observed enrichment for ASD-associated genomic mechanisms particularly in regions that differed between increasers and decreasers. Variability in cortical volume and surface area was associated with enrichment for genes transcriptionally downregulated in ASD postmortem cortical tissue. Previous studies have linked these genes (through GO enrichment analysis) to synaptic proteins, but this annotation may be incomplete. Specifically, many of these genes are also integrally involved in early processes associated with cell proliferation or neurogenesis (51–53)—that is, they have pleiotropic roles at different points in development that may affect mechanisms relevant to cortical volume and surface area and also the synapse. Moreover, neuroanatomical atypicality was correlated with genetic variation associated with ASD. Specifically, in regions differing between increasers and decreasers, greater deviation from the typical developmental trajectory of cortical thickness was correlated with a greater autism PGS restricted to the genes expressed here. This finding builds on previous reports associating (atypical) cortical thickness with genetic variation in autistic children (17) by demonstrating a relationship between (atypical) neuroanatomy and genes not only globally but regionally, that is, in regions relevant to outcome, and not only in children but across age. Taken together, our findings suggest that particular aspects of adaptive outcome–related neuroanatomical variability in ASD may be accompanied by specific biological mechanisms relevant to these phenotypes.
In summary, we identified developmental differences in neuroanatomy and associated genetic factors that are linked to variation in subsequent adaptive behavioral change in ASD. If validated, our findings may enable the future stratification of patient groups, for example, to help target those more likely to have relatively poor outcomes, and enhance efforts to develop better targeted (personalized medicine) interventions in ASD.
1. : Diagnostic and Statistical Manual of Mental Disorders, 5th ed. Washington, DC, American Psychiatric Association, 2013Crossref, Google Scholar
2. : Autism spectrum disorder. Nat Rev Dis Primers 2020; 6:5Crossref, Medline, Google Scholar
3. : Investigating the factors underlying adaptive functioning in autism in the EU-AIMS Longitudinal European Autism Project. Autism Res 2019; 12:645–657Crossref, Medline, Google Scholar
4. : Mid-life social outcomes for a population-based sample of adults with ASD. Autism Res 2018; 11:142–152Crossref, Medline, Google Scholar
5. : Measuring social communication behaviors as a treatment endpoint in individuals with autism spectrum disorder. Autism 2015; 19:622–636Crossref, Medline, Google Scholar
6. : Efficacy of low-dose buspirone for restricted and repetitive behavior in young children with autism spectrum disorder: a randomized trial. J Pediatr 2016; 170:45–53.e1-4Crossref, Medline, Google Scholar
7. : Arbaclofen in children and adolescents with autism spectrum disorder: a randomized, controlled, phase 2 trial. Neuropsychopharmacology 2017; 42:1390–1398Crossref, Medline, Google Scholar
8. : Continuity and change from early childhood to adolescence in autism. J Child Psychol Psychiatry 2005; 46:401–408Crossref, Medline, Google Scholar
9. : Regression, developmental trajectory, and associated problems in disorders in the autism spectrum: the SNAP study. J Autism Dev Disord 2008; 38:1827–1836Crossref, Medline, Google Scholar
10. : The role of adaptive behavior in autism spectrum disorders: implications for functional outcome. J Autism Dev Disord 2011; 41:1007–1018Crossref, Medline, Google Scholar
11. : Outcome and prognostic factors in infantile autism and similar conditions: a population-based study of 46 cases followed through puberty. J Autism Dev Disord 1987; 17:273–287Crossref, Medline, Google Scholar
12. : Cognitive and language skills in adults with autism: a 40-year follow-up. J Child Psychol Psychiatry 2014; 55:49–58Crossref, Medline, Google Scholar
13. : Resting-state functional connectivity predicts longitudinal change in autistic traits and adaptive functioning in autism. Proc Natl Acad Sci U S A 2015; 112:E6699–E6706Crossref, Medline, Google Scholar
14. : Amygdala enlargement in toddlers with autism related to severity of social and communication impairments. Biol Psychiatry 2009; 66:942–949Crossref, Medline, Google Scholar
15. : Neural correlates of communication skill and symptom severity in autism: a voxel-based morphometry study. Res Autism Spect Dis 2009; 3:444–454Crossref, Google Scholar
16. : A diffusion tensor imaging study in children with ADHD, autism spectrum disorder, OCD, and matched controls: distinct and non-distinct white matter disruption and dimensional brain-behavior relationships. Am J Psychiatry 2016; 173:1213–1222Link, Google Scholar
17. : Synaptic and transcriptionally downregulated genes are associated with cortical thickness differences in autism. Mol Psychiatry 2019; 24:1053–1064Crossref, Medline, Google Scholar
18. : Interindividual differences in cortical thickness and their genomic underpinnings in autism spectrum disorder. Am J Psychiatry
19. : The EU-AIMS Longitudinal European Autism Project (LEAP): design and methodologies to identify and validate stratification biomarkers for autism spectrum disorders. Mol Autism 2017; 8:24Crossref, Medline, Google Scholar
20. : Vineland II: Vineland Adaptive Behavior Scales. Circle Pines, Minn, American Guidance Service Publishing, 2005Google Scholar
21. : The EU-AIMS Longitudinal European Autism Project (LEAP): clinical characterisation. Mol Autism 2017; 8:27Crossref, Medline, Google Scholar
22. : Adaptive behavior in autism: minimal clinically important differences on the Vineland–II. Autism Res 2018; 11:270–283Crossref, Medline, Google Scholar
23. : The effect of age, diagnosis, and their interaction on vertex-based measures of cortical thickness and surface area in autism spectrum disorder. J Neural Transm (Vienna) 2014; 121:1157–1170Crossref, Medline, Google Scholar
24. : Variation in longitudinal trajectories of regional brain volumes of healthy men and women (ages 10–85 years) measured with atlas-based parcellation of MRI. Neuroimage 2013; 65:176–193Crossref, Medline, Google Scholar
25. : Detecting changes in nonisotropic images. Hum Brain Mapp 1999; 8:98–101Crossref, Medline, Google Scholar
26. : Specification of cerebral cortical areas. Science 1988; 241:170–176Crossref, Medline, Google Scholar
27. : Early brain overgrowth in autism associated with an increase in cortical surface area before age 2 years. Arch Gen Psychiatry 2011; 68:467–476Crossref, Medline, Google Scholar
28. : The Autism Diagnostic Observation Schedule–Generic: a standard measure of social and communication deficits associated with the spectrum of autism. J Autism Dev Disord 2000; 30:205–223Crossref, Medline, Google Scholar
29. : Social Responsiveness Scale: SRS-2, 2nd ed. Torrance, Calif, Western Psychological Services, 2012, p. 106Google Scholar
30. : An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 2012; 489:391–399Crossref, Medline, Google Scholar
31. : Correlated gene expression supports synchronous activity in brain networks. Science 2015; 348:1241–1244Crossref, Medline, Google Scholar
32. : Autism Diagnostic Observation Schedule: a standardized observation of communicative and social behavior. J Autism Dev Disord 1989; 19:185–212Crossref, Medline, Google Scholar
33. : Autism diagnostic interview-revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord 1994; 24:659–685Crossref, Medline, Google Scholar
34. : Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007; 8:118–127Crossref, Medline, Google Scholar
35. : Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 2018; 362:eaat8127Crossref, Medline, Google Scholar
36. : Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 2016; 540:423–427Crossref, Medline, Google Scholar
37. : Neural correlates of social cognition in naturalistic settings: a model-free analysis approach. Neuroimage 2010; 49:894–904Crossref, Medline, Google Scholar
38. : Anatomical differences in the mirror neuron system and social cognition network in autism. Cereb Cortex 2006; 16:1276–1282Crossref, Medline, Google Scholar
39. : Motor learning in individuals with autism spectrum disorder: activation in superior parietal lobule related to learning and repetitive behaviors. Autism Res 2015; 8:38–51Crossref, Medline, Google Scholar
40. : Insula response and connectivity during social and non-social attention in children with autism. Soc Cogn Affect Neurosci 2016; 11:433–444Crossref, Medline, Google Scholar
41. : Saliency, switching, attention, and control: a network model of insula function. Brain Struct Funct 2010; 214:655–667Crossref, Medline, Google Scholar
42. : Decreased structural connectivity and resting-state brain activity in the lateral occipital cortex is associated with social communication deficits in boys with autism spectrum disorder. Neuroimage 2019; 190:205–212Crossref, Medline, Google Scholar
43. : Neocerebellar contributions to social perception in adolescents with autism spectrum disorder. Dev Cogn Neurosci 2014; 10:77–92Crossref, Medline, Google Scholar
44. : Dynamic mapping of human cortical development during childhood through early adulthood. Proc Natl Acad Sci U S A 2004; 101:8174–8179Crossref, Medline, Google Scholar
45. : Regional gray matter volumetric changes in autism associated with social and repetitive behavior symptoms. BMC Psychiatry 2006; 6:56Crossref, Medline, Google Scholar
46. : Reduced gray matter volume of pars opercularis is associated with impaired social communication in high-functioning autism spectrum disorders. Biol Psychiatry 2010; 68:1141–1147Crossref, Medline, Google Scholar
47. : Pax6 controls cerebral cortical cell number by regulating exit from the cell cycle and specifies cortical cell identity by a cell autonomous mechanism. Dev Biol 2007; 302:50–65Crossref, Medline, Google Scholar
48. : Expression of the transcription factor, tailless, is required for formation of superficial cortical layers. Cereb Cortex 2003; 13:921–931Crossref, Medline, Google Scholar
49. : Id4 regulates neural progenitor proliferation and differentiation in vivo. Development 2004; 131:5441–5448Crossref, Medline, Google Scholar
50. : Spatial gene expression analysis of neuroanatomical differences in mouse models. Neuroimage 2017; 163:220–230Crossref, Medline, Google Scholar
51. : Genetics studies indicate that neural induction and early neuronal maturation are disturbed in autism. Front Cell Neurosci 2014; 8:397Crossref, Medline, Google Scholar
52. : Maternal immune activation dysregulation of the fetal brain transcriptome and relevance to the pathophysiology of autism spectrum disorder. Mol Psychiatry 2018; 23:1001–1013Crossref, Medline, Google Scholar
53. : The ASD living biology: from cell proliferation to clinical phenotype. Mol Psychiatry 2019; 24:88–107Crossref, Medline, Google Scholar



