Interindividual Differences in Cortical Thickness and Their Genomic Underpinnings in Autism Spectrum Disorder
Abstract
Objective:
Autism spectrum disorder (ASD) is accompanied by highly individualized neuroanatomical deviations that potentially map onto distinct genotypes and clinical phenotypes. This study aimed to link differences in brain anatomy to specific biological pathways to pave the way toward targeted therapeutic interventions.
Methods:
The authors examined neurodevelopmental differences in cortical thickness and their genomic underpinnings in a large and clinically diverse sample of 360 individuals with ASD and 279 typically developing control subjects (ages 6–30 years) within the EU-AIMS Longitudinal European Autism Project (LEAP). The authors also examined neurodevelopmental differences and their potential pathophysiological mechanisms between clinical ASD subgroups that differed in the severity and pattern of sensory features.
Results:
In addition to significant between-group differences in “core” ASD brain regions (i.e., fronto-temporal and cingulate regions), individuals with ASD manifested as neuroanatomical outliers within the neurotypical cortical thickness range in a wider neural system, which was enriched for genes known to be implicated in ASD on the genetic and/or transcriptomic level. Within these regions, the individuals’ total (i.e., accumulated) degree of neuroanatomical atypicality was significantly correlated with higher polygenic scores for ASD and other psychiatric conditions, and it scaled with measures of symptom severity. Differences in cortical thickness deviations were also associated with distinct sensory subgroups, especially in brain regions expressing genes involved in excitatory rather than inhibitory neurotransmission.
Conclusions:
The study findings corroborate the link between macroscopic differences in brain anatomy and the molecular mechanisms underpinning heterogeneity in ASD, and provide future targets for stratification and subtyping.
There is increasing recognition that major psychiatric conditions are highly heterogeneous at both the causative and phenotypic levels (1). Assessing this heterogeneity is crucial to allowing more precise inferences about the etiological mechanisms underpinning these conditions and to pave the way toward better targeted (i.e., personalized) therapeutic interventions. This particularly applies to autism spectrum disorder (ASD), which is a common neurodevelopmental condition that is clinically characterized by atypical social communication and interaction, the presence of rigid, repetitive, and stereotyped behaviors, and atypical sensory processing (2). These core ASD symptoms emerge alongside atypical anatomical development of several neurocognitive systems (3, 4). Regional neuroanatomical differences in ASD, however, typically have small effect sizes (5) and reflect highly variable patterns of neurodevelopmental deviations across individuals (6). This makes it inherently difficult to link atypical brain structure to molecular and pathophysiological mechanisms.
Few studies to date have explored the genomic mechanisms underpinning atypical neurodevelopment in ASD. Notably, a recent study by Romero-Garcia et al. (7) demonstrated that differences in cortical thickness during childhood are robustly associated with genes involved in synaptic transmission pathways that are known to be downregulated in the postmortem ASD cortex (7, 8). The study represents an important step toward closing the gap between molecular and macroscopic pathology in ASD, and it highlights the potential of in vivo neuroimaging markers for patient stratification and subtyping. However, the study examined only a narrow range of individuals across the autism spectrum, namely, 10-year-olds with an average full-scale IQ. It therefore remains unknown whether or how these findings generalize to the wider autism phenotype, which is highly heterogeneous, not only between individuals meeting diagnostic criteria (9) but also within individuals across development (10).
In this study, we explored interindividual differences in cortical thickness and their potential molecular underpinnings in a large, clinically diverse sample of individuals with ASD and control subjects within the EU-AIMS Longitudinal European Autism Project (LEAP; www.aims-2-trials.eu) (11). This study provides deep phenotypic assessments of and genotypic data for more than 700 individuals, including males and females between the ages of 6 and 30 years, with a wide range of intellectual abilities and varying degrees of symptom severity (9). Rather than exclusively focusing on “core” ASD regions (i.e., regions with a significant between-group difference), we based our investigation on local (i.e., regional) and global (i.e., cortex-level) neuroanatomical deviations from the neurotypical range of cortical thickness in our sample. A similar approach has been used previously to assess neuroanatomical heterogeneity in ASD (6, 12) and for the biologically driven stratification of ASD into putative subtypes (13). Here, we used the technique to identify a wider set of brain regions where being a neuroanatomical outlier significantly has an impact on the probability (i.e., risk) of ASD in our sample, and to link patterns of neuroanatomical variability to ASD symptomatology and genetic risk factors.
More specifically, leveraging the spatial gene expression data of the Allen Human Brain Atlas (14), we tested the hypothesis that brain regions associated with high neuroanatomical variability in ASD express more than expected genes that are enriched for rare (15) or common ASD risk variants (16) and/or are abnormally expressed in ASD (8, 17, 18). Moreover, using a virtual histology approach (19), we probed the potential molecular mechanisms underpinning clinical heterogeneity in ASD via patterns of neurodevelopmental deviations that differed between distinct sensory ASD subgroups (20). We focused on sensory symptoms, as these have previously been highlighted as promising candidates in parsing heterogeneity in ASD (21). Moreover, sensory features have etiologically been linked to an imbalance between excitation and inhibition (22, 23), which provides a reasonable conceptual framework for hypothesis testing.
Methods
Participants
This study utilized data provided by the EU-AIMS LEAP, a multicenter transdisciplinary study on stratification biomarkers for ASD (www.eu-aims.eu). A comprehensive description of the sample has been published elsewhere (9). In brief, the total sample for which usable structural MRI data were available included 360 individuals with ASD and 279 control subjects, split into 254 typically developing participants and 25 individuals with mild intellectual disability (14 males and 11 females; defined by full-scale IQ between 50 and 74), between the ages of 6 and 30 years (Table 1; see also Figure S1 in the online supplement). A full list of inclusion and exclusion criteria, clinical assessments, and medication status is provided in the online supplement. An independent ethics committee approved the study, and written informed consent was obtained for all participants.
| Variable | Typically Developing Group (N=279) | ASD Group (N=360) | Analysis | ||||
|---|---|---|---|---|---|---|---|
| N | % | N | % | χ2 | df | p | |
| Sex | 4.45 | 1 | <0.03 | ||||
| Male | 178 | 63.8 | 259 | 71.9 | |||
| Female | 101 | 36.2 | 101 | 28.1 | |||
| Mean | SD | Mean | SD | t | p | ||
| Age (years) | 17.33 | 5.91 | 17.51 | 5.51 | –0.40 | 576 | 0.69 |
| Full-scale IQ | 104.79 | 18.25 | 98.86 | 19.72 | 3.93 | 617 | <0.01 |
| ADI-R scores | |||||||
| Social interaction | 16.68 | 6.69 | |||||
| Communication | 13.22 | 5.63 | |||||
| Restricted and repetitive behaviors | 4.29 | 2.67 | |||||
| ADOS scores | |||||||
| Total | 6.07 | 2.64 | |||||
| Social affect | 6.77 | 2.40 | |||||
| Restricted and repetitive behaviors | 4.70 | 2.62 | |||||
| Total brain volume (liters) | 1.19 | 0.13 | 1.19 | 0.13 | –0.15 | 603 | 0.88 |
| Mean cortical thickness (mm) | 2.67 | 0.11 | 2.68 | 0.13 | –0.78 | 623 | 0.43 |
TABLE 1. Characteristics of participants with autism spectrum disorder (ASD) and typically developing control subjectsa
MRI Data Acquisition
All participants underwent MR imaging in 3-T scanners, at six different sites (University of Cambridge and King’s College London, U.K.; Mannheim University, Germany; Radboud University and Utrecht University, the Netherlands; Rome University, Italy). High-resolution structural T1-weighted volumetric images were acquired with full head coverage, at 1.2-mm thickness with 1.2 × 1.2-mm in-plane resolution (see Table S2 the online supplement for details).
Cortical Surface Reconstructions Using FreeSurfer
Usable structural MRI data were initially available for 709 individuals in the LEAP sample. FreeSurfer, version 6.0.0 (http://surfer.nmr.mgh.harvard.edu/) was used to derive models of the cortical surface for each T1-weighted image. These well-validated and fully automated procedures have been described extensively elsewhere (24, 25). Each reconstructed surface underwent strict quality assessments (see the online supplement), resulting in a final sample of 639. We examined measures of cortical thickness, which represent the closest distance from the outer (i.e., pial) to the inner (i.e., white) matter boundary at each vertex on the tessellated surface (26), smoothed using a 15-mm kernel (see the online supplement for details). For each participant, we also computed mean cortical thickness across the cortex (CT0).
Surface-Based Statistical Analyses of Cortical Thickness
Statistical analyses were conducted using the SurfStat toolbox (http://www.math.mcgill.ca/keith/surfstat) for MATLAB, version R2017b (https://www.mathworks.com) and R for Statistical Computing (www.r-project.org). Vertex-wise between-group differences in cortical thickness (Y) were examined by regression of a general linear model with 1) diagnostic group, sex, and acquisition site as fixed-effect factors (see the online supplement for site effects), and 2) linear and quadratic age, full-scale IQ (FSIQ), and CT0 as continuous covariates:
 is the residual error at vertex i. Between-group differences were estimated from the coefficient β1, normalized by the standard error. All continuous covariates were mean centered across groups. Corrections for multiple comparisons were performed using random-field-theory-based cluster analysis for nonisotropic images with a cluster-based significance threshold (pclust) of 0.05 (two-tailed [27]). Effect sizes associated with each model term were assessed using Cohen’s f, where values of 0.1, 0.25, and 0.4 indicate small, medium, and large effects, respectively. At each vertex, we also used a Levene’s test to assess between-group differences in cortical thickness variability at a false discovery rate (FDR) corrected p value (padj) <0.05. Brain-behavior correlations were examined using Pearson’s r.
is the residual error at vertex i. Between-group differences were estimated from the coefficient β1, normalized by the standard error. All continuous covariates were mean centered across groups. Corrections for multiple comparisons were performed using random-field-theory-based cluster analysis for nonisotropic images with a cluster-based significance threshold (pclust) of 0.05 (two-tailed [27]). Effect sizes associated with each model term were assessed using Cohen’s f, where values of 0.1, 0.25, and 0.4 indicate small, medium, and large effects, respectively. At each vertex, we also used a Levene’s test to assess between-group differences in cortical thickness variability at a false discovery rate (FDR) corrected p value (padj) <0.05. Brain-behavior correlations were examined using Pearson’s r.
To quantify neuroanatomical deviations from the neurotypical cortical thickness range, we fitted a general linear model within the neurotypical control subjects without intellectual disability that included age, sex, full-scale IQ, site, and CT0 as predictors (X) (see the online supplement for effects of intellectual disability and sex). The model coefficients ( , where TD signifies typically developing) were subsequently utilized to predict cortical thickness for individuals with ASD and control subjects with intellectual disability (
, where TD signifies typically developing) were subsequently utilized to predict cortical thickness for individuals with ASD and control subjects with intellectual disability ( ). The resulting residuals (
). The resulting residuals ( ) were centered and scaled based on the neurotypical cortical thickness distribution, thus expressing all data in unit standard deviations of the predicted neurotypical mean (zres). Based on these deviations, we identified vertex-level outliers defined as cortical thickness values falling outside the neurotypical 90% prediction interval (PI90%) (see the online supplement on PI threshold). This resulted in an n-by-p matrix of either zeros (inside PI90%) or ones (outside PI90%), where n denotes participants and p vertices. Based on the outlier matrix, we computed vertex-wise estimates of the model’s in-sample 1) sensitivity, that is, the probability of being a neuroanatomical outlier given that an individual has ASD (p[PI90%,out|ASD]); 2) specificity, that is, the probability of being inside the neurotypical PI90% for non-ASD individuals (p[PI90%,in|TD]); 3) positive predictive value, that is, the probability of ASD for individuals outside the neurotypical PI90% (p[ASD|PI90%,out]); and 4) negative predictive value, that is, the probability of not having ASD for individuals inside the neurotypical PI90% (p[TD|PI90%,in]). At each vertex, we also compared the sample prevalence of ASD (pretest probability) with the probability of ASD given that an individual falls outside the neurotypical PI90% (posttest probability).
) were centered and scaled based on the neurotypical cortical thickness distribution, thus expressing all data in unit standard deviations of the predicted neurotypical mean (zres). Based on these deviations, we identified vertex-level outliers defined as cortical thickness values falling outside the neurotypical 90% prediction interval (PI90%) (see the online supplement on PI threshold). This resulted in an n-by-p matrix of either zeros (inside PI90%) or ones (outside PI90%), where n denotes participants and p vertices. Based on the outlier matrix, we computed vertex-wise estimates of the model’s in-sample 1) sensitivity, that is, the probability of being a neuroanatomical outlier given that an individual has ASD (p[PI90%,out|ASD]); 2) specificity, that is, the probability of being inside the neurotypical PI90% for non-ASD individuals (p[PI90%,in|TD]); 3) positive predictive value, that is, the probability of ASD for individuals outside the neurotypical PI90% (p[ASD|PI90%,out]); and 4) negative predictive value, that is, the probability of not having ASD for individuals inside the neurotypical PI90% (p[TD|PI90%,in]). At each vertex, we also compared the sample prevalence of ASD (pretest probability) with the probability of ASD given that an individual falls outside the neurotypical PI90% (posttest probability).
The individuals’ accumulated degree of neuroanatomical atypicality was summarized in a subject-level total neuroanatomical atypicality index (tAIs), which indicates the percentage of vertices outside the neurotypical PI90% per individual. The tAIs values were computed based on 1) all vertices on the cortical surface, and 2) within an outlier mask that included only vertices with a significant chi-square enrichment of ASD outside the neurotypical PI90% (i.e., p[ASD|PI90%,out] > p[ASD|PI90%,in]). The tAIs values were subsequently used for the comparison between groups using a t test for independent samples (p<0.05) and for the prediction of diagnostic categories using a logistic regression model. Moreover, using Pearson’s r, we examined the global impact of neuroanatomical deviations on ASD symptomatology across DSM-5 symptom domains. Last, we tested the hypothesis that the individual’s neuroanatomical load is significantly correlated with the polygenic risk for ASD and other psychiatric conditions.
Genetic Analyses
Genome-wide polygenic scores (PGSgenome) for ASD (16) and a variety of other psychiatric conditions and phenotypic traits (e.g., attention deficit hyperactivity disorder [ADHD] [28], schizophrenia [29], depression [30], and epilepsy [31]) were derived as outlined in the online supplement. In addition, using the PRSet function in PRSice-2 (32), we derived gene set–based polygenic scores (PGSset) for ASD (using the genome-wide association study [GWAS] summary statistics described in reference 16) across gene sets that are highly expressed within brain areas highlighted by our neuroimaging findings (see the online supplement for details). To this aim, we performed a gene expression decoding analysis within Neurosynth and NeuroVault (33) to identify genes whose spatial expression patterns resemble our neuroimaging findings (see the online supplement for details). In brief, this analysis utilizes the gene expression data from the Allen Human Brain Atlas (14) to statistically assess the spatial correlation between our neuroimaging maps (t-map, Cohen’s f-map, chi-square outlier maps) and the patterns of expression for each of 20,787 protein coding genes. To do so, the six donor brains of the Allen atlas are initially coregistered with the neuroimaging data to bring normalized gene expression values into transcriptomic alignment with the FreeSurfer surface overlays (see Figure S20 in the online supplement). A linear model is then constructed for each donor brain, where the slopes encode the spatial correlation between each gene’s expression pattern and the values contained in the statistical maps at each probe (i.e., sampling site). The slopes are then subjected to a one-sample t test to identify genes whose expression patterns are consistently (i.e., across donor brains) highly similar to the imaging maps.
The resulting gene lists for each statistical map were thresholded at p<0.01. We chose this liberal threshold because this analysis did not constitute a hypothesis test perse, but rather a selection step to provide a list of candidate genes. This list was subsequently tested for enrichment with genes previously implicated in ASD in genetic and transcriptomic studies. At the genetic level, this included ASD risk genes with de novo and rare variants (15) and GWAS-significant ASD risk genes with common variants (16). At the level of differential gene expression, we tested gene lists that are 1) differentially expressed (i.e., upregulated or downregulated) in postmortem cortical tissue (17) and in specific neuronal cell types in ASD (18), and 2) genes of differentially expressed coregulated modules in ASD (8, 34). We also included the ASD gene list compiled by the SFARI database (categories S, 1, 2, and 3, downloaded in November 2020 from https://gene.sfari.org/). Notably, these gene sets are partially overlapping (see Figure S22 in the online supplement for number of total and intersecting genes).
In addition, we tested for an enrichment of genes underpinning typical brain development via the human brain transcriptome data set provided by Kang et al. (35), which covers transcriptome profiles of 16 different brain regions from embryonic development to late adulthood. For this purpose, two-dimensional heat maps representing the time course of gene expression across different brain regions were created based on the module “eigengene,” as implemented in the MAGMA pipeline (https://github.com/SheenYo/MAGNET) (36). All enrichment testing was performed using the GeneOverlap package in R (10.18129/B9.bioc.GeneOverlap), which generated enrichment odds ratios, hypergeometric p values, and FDR-corrected p values (padj). Only comparisons with padj values <0.05 were interpreted further. To establish the relative impact of differentially expressed genes (DEGs) and ASD risk genes on the individuals’ degree of neurodevelopmental perturbation, we predicted tAIs values using set-based PGS across 1) gene sets with atypical expression that were significantly enriched in the chi-square outlier map (PGSDGE, where DGE signifies genes with differential gene expression in ASD), and 2) ASD risk genes with common and de novo variants (PGSrisk) (15, 16) (see Table S4 in the online supplement for details).
Neuroanatomical Differences and Genetic Underpinnings of Clinical ASD Subgroups
To relate distinct clinical ASD phenotypes to different patterns of neuroanatomical deviations, we stratified individuals with ASD based on the severity and profile of sensory symptoms. The subgroups were originally derived by using a factor mixture modeling approach (20) across questionnaire items of the Short Sensory Profile (37) (see Table S5 in the online supplement for summary scores), which resulted in three groups: “sensory low” (N=209), “sensory moderate” (N=37), and “sensory severe” (N=18) individuals in our ASD sample. At each vertex, an F-test for the main effect of sensory subgroup was performed based on the individuals’ standardized cortical thickness deviations (zres) using a random-field-theory-corrected p value <0.05. Given the link between atypical sensory processing and excitation-inhibition imbalance (22), we employed a virtual histology approach to relate regional differences between ASD subgroups to those in cell-specific gene expression (19). To this end, a gene expression decoding analysis of the resulting F-map was performed as outlined above. The list of significant decoded genes was subsequently tested for cell-type enrichment based on the Single-Cell Transcriptomic Atlas of Human Neocortical Development During Midgestation (38), which, among other cell-type specific genes, allowed us to test for an enrichment of genes representing different subtypes of excitatory and inhibitory cells.
Results
Overall, the ASD group and the typically developing control group were matched for age, total brain volume, and mean cortical thickness (Table 1). However, the ASD group had a significantly lower mean full-scale IQ (mean=98.8, SD=20) than the control group (mean=104.8, SD=18). Our sample included more males with ASD than male control subjects, and more females than males.
Vertex-Wise Between-Group Differences in Cortical Thickness
Individuals with ASD had increased cortical thickness relative to control subjects in the left and right anterior cingulate cortex (ACC; approximate Brodmann areas [BA] 24/33), the left and right anterior temporal lobes (BA 20/21/22/38/41/42), the left lingual gyrus (BA 18/19), and the right posterior cingulate cortex (BA 23/31). By contrast, decreased cortical thickness in the ASD group was observed in the left dorsolateral prefrontal cortex (DLPFC; BA 4/6/8/9) and precentral gyrus (BA 4/6), the right parahippocampal and fusiform gyrus (BA 18/19/34/37), the left temporal pole (BA 20/38), and the right pre- and postcentral gyrus (BA 4/6) (Figure 1A,B; see also Table S2 in the online supplement). In these brain regions, measures of cortical thickness within the ASD group were also significantly correlated with measures of symptom severity across DSM-5 symptom domains (Figure 1G,H).
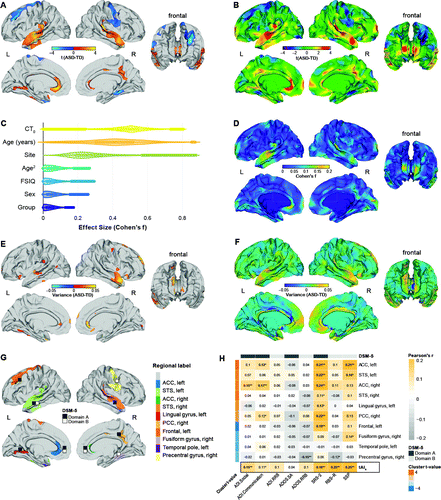
FIGURE 1. Vertex-wise between-group differences in cortical thickness among individuals with autism spectrum disorder (ASD) and control subjectsa
a Panel A shows clusters with significantly increased (orange to yellow) and decreased (blue to cyan) cortical thickness in the ASD group relative to the control group (random-field-theory-based cluster corrected p<0.05, two-tailed). TD=typically developing control subjects. Panel B shows the t test statistic for the contrast ASD versus control (unthresholded). Panel C shows effect sizes associated with individual model terms across the cortex. CT0=mean cortical thickness; FSIQ=full-scale IQ. Panel D shows the spatially distributed pattern of effects (Cohen’s f) for the main effect of group. Panel E shows vertices with significantly increased variance in cortical thickness in the ASD group relative to the control group resulting from a Levene’s test of homogeneity of variances (false discovery rate [FDR] corrected p<0.05). Panel F shows absolute vertex-level differences in cortical thickness variability between groups. Panel G shows clusters with significant bivariate Pearson correlation coefficients between cortical thickness and measures of symptom severity subdivided into domains A and B of DSM-5 within the ASD group. ACC=anterior cingulate cortex; PCC=posterior cingulate cortex; STS=superior temporal sulcus. Panel H shows brain-behavior correlations between clusters with significant differences in cortical thickness and measures of symptom severity subdivided into DSM-5 domain A and B symptoms within the ASD group. Correlation coefficients in boldface survived an FDR-corrected p value threshold of 0.05. ADI=Autism Diagnostic Interview–Revised; ADOS.SA/ADOS.RRB=Autism Diagnostic Observation Schedule calibrated severity score for social affect (SA) and restricted and repetitive behaviors (RRB); RRB=repetitive and restricted behaviors; RBS-R=Repetitive Behaviors Scale–Revised; SRS-2=Social Responsiveness Scale–2; SSP=Short Sensory Profile (reverse-scored so that larger values indicate more severe symptoms); tAIs=subject-level total neuroanatomical abnormality index.
*p<0.05. **p<0.001.
Vertex-level effect sizes (Cohen’s f) for the main effect of group were small overall, ranging from 0 to 0.174 across the cortex (mean=0.042, SD=0.032) (Figure 1C), which is consistent with small mean differences and significantly increased cortical thickness variability within the ASD group (Figure 1E,F). The largest effect sizes were observed in the left and right temporal lobes and the ACC, as well as in anterior medial frontal and occipital regions (Figure 1D). Effect sizes for the main effect of group were also relatively low compared with the effects of other model terms, such as total brain volume (mean=0.477, SD=0.012), age (mean=0.405, SD=0.168), and acquisition site (mean=0.279, SD=0.130) (Figure 1C), each of which displayed a unique pattern of spatial variability (see Figure S2 in the online supplement).
Vertex-Level Deviations From the Neurotypical Distribution of Cortical Thickness
Based on the neurotypical PI90% in our sample, we initially identified vertex-level neuroanatomical outliers. At any given vertex, maximally 22.2% of individuals with ASD fell outside the neurotypical range (mean=11.2%, SD=2.5% across vertices), with the highest proportion of outliers being observed in the left and right temporal and medial prefrontal regions (Figure 2A; see Figure S26 in the online supplement for standard residual error). Moreover, there was considerable interindividual variability with regard to the pattern and direction of neuroanatomical deviations observed, with some individuals displaying predominantly positive deflections, negative deflections, or a mixture of both (see Figure S6 in the online supplement).
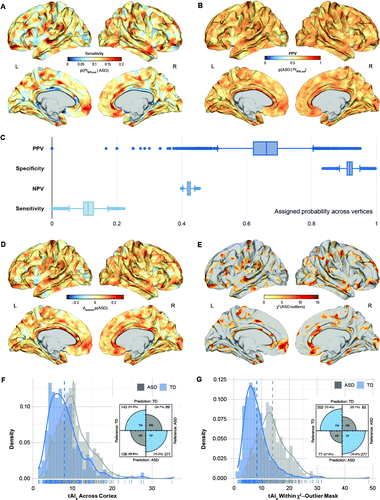
FIGURE 2. Vertex-level outlier statistics from the neurotypical range of cortical thickness predicted by age, sex, full-scale IQ, site, and mean cortical thickness across the cortex among individuals with autism spectrum disorder (ASD) and control subjectsa
a Panel A shows the probability of falling outside the neurotypical 90% prediction interval (PI90%) given that an individual has ASD. This equals the proportion of individuals with ASD falling outside the neurotypical PI90% at each vertex (i.e., sensitivity). Panel B shows the prevalence (i.e., probability) of ASD among all individuals outside the neurotypical PI90%. This equals the positive predictive value (PPV) of the model. Panel C shows the sensitivity (true positive rate), specificity (true negative rate), positive predictive value (PPV), and negative predictive value (NPV) of the neurotypical model across all vertices on the cortical surface. Panel D shows increases (yellow to red) and decreases (cyan to blue) in the posttest probability of ASD based on a pretest probability that equals prevalence of ASD in our sample. Panel E shows vertices with a significant chi-square enrichment of individuals with ASD outside the neurotypical PI90%. Panel F shows between-group differences in the individuals’ total degree of neuroanatomical abnormality (tAIs), quantified by the percentage of vertices outside the neurotypical PI90% across the cortex, and panel G shows between-group differences quantified by the percentage of vertices within the chi-square enrichment mask. Subplots display the results of the logistic regression analysis, predicting diagnostic categories based on the individuals’ tAIs. TD=typically developing control subjects; FN=false negative rate; FP=false positive rate; TN=true negative rate; TP=true positive rate. Note that FN, FP, TN, and TP represent sample estimates rather than quantifying generalization performance.
Among all individuals outside the neurotypical PI90%, the probability of ASD was very high (i.e., positive predictive value), with some vertices displaying a positive predictive value as high as 95% (mean=65.6%, SD=5.8%) (Figure 2B). More specifically, taking into account the sensitivity, specificity, and ASD sample prevalence (0.56), we found that in some brain regions, being a neuroanatomical outlier increased the risk of a diagnosis of ASD by as much as 38.2% (mean=7.1%, SD=5.95) (Figure 2D). Consequently, there were a number of vertices with a significant chi-square enrichment of ASD outside the neurotypical PI90%, which highlights brain regions where 1) the risk of ASD is significantly modulated by being a neuroanatomical outlier, and 2) individuals with ASD have a higher outlier probability than control subjects (see chi-square outlier map in Figure 2E).
Individuals with ASD also had a significantly larger total degree of neuroanatomical abnormality (tAIs) across the cortex (t=−7.123, df=609, p<0.001) and within the chi-square map of significant neuroanatomical outliers (t=−14.316, df=634, p<0.001) (Figure 2F,G). This difference was sufficient to separate groups at an overall accuracy of 74.96% (64.79% across the cortex) in our sample (odds ratio=1.292, βtAI=0.256, p<0.001), with a sensitivity of 76.94% and a specificity of 72.40% (positive predictive value=78.25%, negative predictive value=70.88%, where the negative predictive value indicates the probability of not having ASD for individuals inside the neurotypical PI90%). Notably, 84% of the control subjects with intellectual disability were identified as neuroanatomical outliers. Thus, while our tAIs model seems highly sensitive to neurodevelopmental deviations, the detected outliers are not specific to ASD but may include other conditions associated with atypical neurodevelopment. Measures of tAIs were also significantly correlated with symptom severity in the social domain of the Autism Diagnostic Interview–Revised (r=0.142, padj<0.05), the Social Responsiveness Scale–2 (r=0.18, padj<0.05), the Repetitive Behaviors Scale–Revised (r=0.25, padj<0.05), and the Short Sensory Profile (r=0.25, padj<0.05).
Gene Expression Decoding and Enrichment Analyses
To link cortical thickness variability to etiological mechanisms, we utilized the Allen Human Brain Atlas to identify genes with a spatial pattern of expression resembling our imaging maps. This resulted in sets of 546, 408, and 662 significant genes, respectively, in the t-map of between-group differences, the Cohen’s f-map of statistical effects, and the chi-square map of neuroanatomical outliers (p<0.01) (Figure 3A). Within these gene sets, we found an enrichment for genes known to be associated with ASD, and particularly for genes with differential gene expression (DGE) during childhood and adolescence (Figure 3B). More specifically, in the Cohen’s f-map and the chi-square map, we observed a significant enrichment for gene sets that are upregulated in ASD, namely, M16.up (34), CTX.M9.up (8), CTX.M19.up (8), ASD.DEGs.up (17), and CTX.M20.up (8) (padj<0.05). These gene sets have previously been linked to Gene Ontology terms representing immune/inflammatory response and the development/regulation of cell differentiation (8, 17, 34). In contrast, the t-map of significant between-group differences showed a high expression of genes that are downregulated in ASD, for example, M12.down (34) and ASD.DEGs.down (17), which represent genes underlying synaptic functioning and transmembrane transporter activity. There was no significant overrepresentation of ASD risk genes representing common (15) or rare and de novo variants (16).
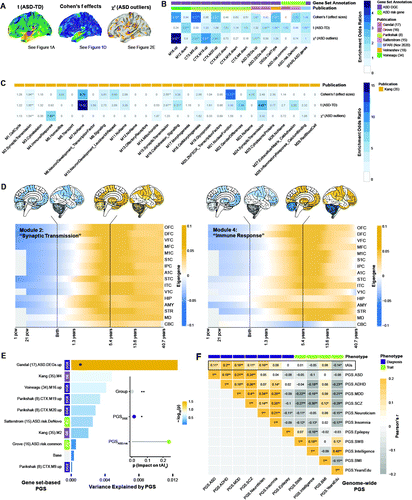
FIGURE 3. Genomic underpinnings of neurodevelopmental deviations in cortical thickness in autism spectrum disorder (ASD)a
a Panel A shows the t-map of statistical between-group differences in cortical thickness (see Figure 1A), the Cohen’s f effect size map associated with the main effect of group (see Figure 1D), and the chi-square map of neuroanatomical ASD outliers (see Figure 2E). TD=typically developing control subjects. Panel B shows significant odds ratios at a false discovery rate (FDR) corrected p threshold of 0.05 resulting from the gene set enrichment analyses for genes expressed in the different output maps. Gene sets were subdivided into sets with differential gene expression in ASD and sets representing ASD risk genes that contain either common variants (ASD.risk.common) or rare de novo variants (ASD.risk.DeNovo). Gene sets are annotated and labeled based on their original publication. CTX=cortex; DEG=differentially expressed gene; down=down-regulated expression in ASD; up=upregulated expression in ASD. Panel C shows set enrichment of genes mediating typical brain development as reported in the spatiotemporal transcriptome data set provided by Kang et al. (35). Set names contain their respective coexpression module label (e.g., M1), followed by their functional description based on their Gene Ontology term enrichment. Panel D shows spatiotemporal expression profiles of brain gene modules significantly enriched in the chi-square outlier map for module 2 (left panel) enriched for genes implicated in synaptic transmission, and for module 4 (right panel) enriched for immune response genes. The x-axis shows the developmental time frame (pcw=postconception weeks) and the y-axis shows the different brain regions: OFC=orbital prefrontal cortex; DFC=dorsolateral prefrontal cortex; VFC=ventrolateral prefrontal cortex; MFC=medial prefrontal cortex; M1C=primary motor (M1) cortex; S1C=primary somatosensory (S1) cortex; IPC=posterior inferior parietal cortex; A1C=primary auditory (A1) cortex; STC=superior temporal cortex; ITC=inferior temporal cortex; V1C=primary visual (V1) cortex; HIP=hippocampus; AMY=amygdala; STR=striatum; MD=mediodorsal nucleus of the thalamus; CBC=cerebellar cortex. Panel E shows gene set–based polygenic scores (PGSs) across gene sets enriched in the chi-square outlier map and across ASD risk genes. Each bar represents the proportion of variance associated with ASD case-control status explained by the PGS within sets. DGE=differential gene expression in ASD; GE=genes expressed in typically developing brain; RG=ASD risk genes with common and de novo variants. The subplot shows the p value associated with the impact of the combined PGS across DGE and GE gene sets (PGSDGE, purple) on the individuals’ total degree of neuroanatomical abnormality (tAIs) relative to the impact of PGS across ASD risk genes (PGSASD.risk, green). Panel F shows Pearson correlations (r) between tAIs and genome-wide PGS for ASD and other neuropsychiatric conditions, as well as general phenotypic traits across groups. ADHD=attention deficit hyperactivity disorder; MDD=major depressive disorder; SCZ=schizophrenia; SWB=subjective well-being; BMI=body mass index; YearsEdu=years of education. PGSs are annotated based on their phenotypical outcome (i.e., clinical diagnosis versus general phenotype trait).
*p<0.05 (FDR-corrected). **p<0.01 (FDR-corrected).
Furthermore, we found an enrichment for gene coexpression modules underpinning typical brain development during the first decades of life (35). We observed a significant enrichment for coexpression modules representing “synaptic transmission” (modules 2, 15, 14) and “cell adhesion signaling” (module M16), which have their highest level of expression during childhood and adolescence (35), and for “nuclear function” module 21 (Figure 3D). In agreement with the analysis of ASD-related genes, the chi-square outlier map also displayed a high expression of genes in “immune response” module 4, which is highly expressed during childhood and adolescence (odds ratio=7.46, padj<0.001). Module 4 was also significantly associated with the ASD genotype in our sample (βM4=0.374, padj<0.01) (see the Gene-Set Analysis section in the online supplement). Odds ratios for all modules and adjusted p values are listed in Figure 3C.
Correlations Between tAIs and Polygenic Score for ASD
Based on the gene set–based PGS (PGSset), we established that genetic variation in differentially expressed genes and coexpression modules enriched in the chi-square outlier map (i.e., PGSDGE) explained a larger percentage of neuroanatomical outliers (tAIs) than PGSset across ASD risk genes with common and de novo variants (i.e., PGSASD.risk) (F=5.99, df=1, p<0.05 and F=1.39, df=1, p=0.23, respectively) when the main effect of group was accounted for (F=142.56, df=1, p<0.001) (Figure 3E). The number of ASD risk genes with common and de novo variants was small overall, however, and included a total of only 1,455 single-nucleotide polymorphisms in our sample (see Table S4 in the online supplement). We therefore also examined the association between genotype and phenotype based on genome-wide PGSs. Measures of tAIs were significantly correlated with the genome-wide risk for ASD (r=0.11, padj<0.05), ADHD (r=0.2, padj<0.01), depression (r=0.16, padj<0.01), schizophrenia (r=0.11, padj<0.05), and neuroticism (r=0.16, padj<0.01) (Figure 3F). Thus, while patterns of cortical thickness are enriched for genes known to be implicated in ASD, composite measures of neuroanatomical atypicality significantly correlated with the risk not only for ASD but also for other neurodevelopmental conditions.
Differences in Neuroanatomical Deviations Across Different Sensory Subgroups
Vertex-level neurodevelopmental cortical thickness deviations from the neurotypical mean (zres) differed significantly between sensory symptom subgroups. There was a significant main effect of subgroup in the right premotor cortex and supplementary motor area (BA 6/8; Fmax=6.239, pclust<0.05), which included the frontal eye fields (Figure 4A,B). Here, individuals with ASD in the “moderate” and sensory “severe” subgroups displayed a significantly larger proportion of negative cortical thickness deviations compared with “sensory low” individuals (χ2=10.131, df=2, p<0.01) (Figure 4C). Neuroanatomical deviations in this region were also significantly correlated with the “movement sensitivity” subdomain of the Short Sensory Profile (r=−0.186, padj<0.01), which was the subdomain that differed the most between subgroups (effect size=2.52) (20) (Figure 4D). Last, we found the F-map for the main effect of sensory subgroup to be enriched for genes expressed in excitatory neurons in the developing cortex (38) (Figure 4E), including an enrichment for migrating excitatory neurons (odds ratio=6.057, padj<0.05) and excitatory neurons in deep layer 1 and 2 (odds ratio=2.43 and 2.13, respectively, padj<0.05) (Figure 4F).
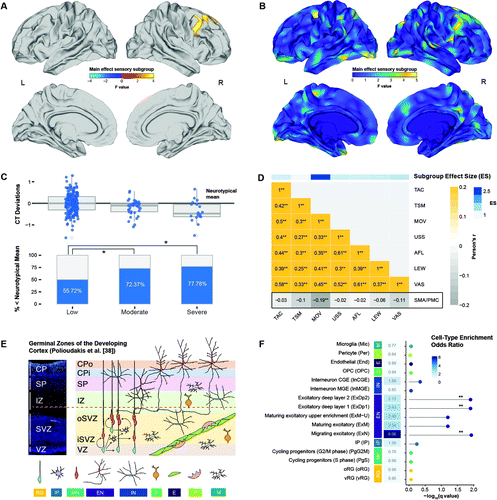
FIGURE 4. Neuroanatomical differences between different ASD sensory subgroupsa
a Panel A shows clusters with a significant main effect of subgroup (random-field-theory-based cluster corrected, p<0.05, two-tailed). Panel B shows F-test statistics for the main effect of subgroup (unthresholded). In panel C, the upper panel shows standardized deviations (i.e., residuals) from the neurotypical distribution of cortical thickness (CT) for the subgroups with low, moderate, and severe sensory symptoms, and the lower panel shows percentage of individuals with deviations falling below the neurotypical mean cortical thickness. Panel D shows Pearson correlation coefficients between neuroanatomical deviations and Short Sensory Profile subdomains at a false discovery rate corrected p threshold of 0.05. TAC=tactile sensitivity; TSM=taste/smell sensitivity; MOV=movement sensitivity; USS=underresponsiveness/seeks sensation; AFL=auditory filtering; LEW=low energy/weak; VAS=visual/auditory sensitivity; SMA/PMC=supplementary motor area/premotor cortex: significant cluster in the supplementary motor area and premotor cortex. The color bar on top shows the subdomain effect size for separating the low from the severe sensory subgroup published in reference 20. Note that the Short Sensory Profile subdomain scores were reverse-scored so that larger values indicate more severe symptoms; a negative correlation thus indicates that more negative neuroanatomical deviations are associated with more severe symptoms. Panel E is a schematic illustration of cell types in germinal zones of the developing cortex, adapted from Polioudakis et al. (38). CP=cortical plate; Cpi=inner cortical plate; CPo=outer cortical plate; SP=subplate; IZ=intermediate zone; SVZ=subventricular zone; iSVZ=inner subventricular zone; oSVZ=outer subventricular zone; VZ=ventricular zone; RG=radial glia; IP=intermediate progenitor; MN=newborn migrating excitatory neuron; EN=excitatory neuron; IN=interneuron; O=oligodendrocyte precursor; E=endothelial cell; P=pericyte; M=microglia. Panel F shows cell-type enrichment odds ratios and associated −log10(q) values for gene sets expressed in the F-map. Cell types are colored and labeled based on Polioudakis et al. (38) (see also Figure 4E). MP=mitotic progenitor; OPC=oligodendrocyte precursors; CGE/MGE=caudal and medial ganglionic eminence-derived interneurons; IP=intermediate progenitors; oRG/vRG=outer and ventricular radial glia.
Discussion
Our study confirms that ASD is accompanied by significant between-group differences in cortical thickness (e.g., in fronto-temporal and cingulate regions) that reflect highly individualized patterns of neurodevelopmental deviations overall. In addition to these core ASD brain areas, we identified a wider spatially distributed network of regions where individuals with ASD and control subjects with intellectual disability manifested as neuroanatomical outliers. This network of regions was enriched for genes known to be upregulated in ASD during childhood and adolescence. Moreover, within this network, the individuals’ total degree of neuroanatomical abnormality was significantly correlated with measures of symptom severity, as well as with the polygenic risk for ASD and other psychiatric conditions. Last, we demonstrate that distinct clinical ASD subgroups display different patterns of neurodevelopmental deviations, which map onto specific cell types in the developing cortex. Our study thus provides novel insights into the genetic and neurobiological mechanisms underpinning heterogeneity in ASD.
Our finding of significant between-group differences in cortical thickness aligns with previous investigations into the neuroanatomical underpinnings of ASD, where both increased and decreased cortical thickness have been reported predominantly in fronto-temporal and fronto-parietal regions (39–41). In these regions, measures of cortical thickness also undergo an abnormal developmental trajectory in ASD (42, 43), with differences being most prominent during childhood and adolescence and diminishing during adulthood (44). The results of most previous studies in samples with Ns <200 have been highly variable with regard to the direction and pattern of cortical thickness differences observed. However, a more consistent picture is now emerging across an increasing number of large-scale investigations (N>500), which typically exclusively report increased cortical thickness in ASD (e.g., 5, 44). Our results thus differ from these latter studies in that we also observed cortical thickness reductions in the DLPFC and precentral gyrus. This discrepancy may be due in part to differences in sample characteristics, preprocessing pipelines, or the composition of the general linear model—for example, we covaried for mean cortical thickness to account for global differences related to brain size (see also Figure S11 in the online supplement). However, the pattern of effect sizes for the main effect of group is remarkably similar across studies (see also reference 12), with the largest effects being reported in the superior temporal sulcus, DLPFC, and posterior cingulate cortex. Thus, taken together, the data from our study and others strongly support the hypothesis that cortical thickness and related aspects of the vertical organization of the cortex (e.g., cortical lamination [45]) are atypical in ASD, and particularly in brain regions that are functionally linked to symptoms and traits (reviewed in reference 4).
Individuals with ASD also manifested as neuroanatomical outliers in a number of brain regions that were not highlighted by the main effect of group, for example, medial prefrontal regions, where falling outside the neurotypical PI90% was associated with increased ASD risk in our sample. Within these brain regions, the pattern and degree (i.e., percentage) of vertex-level outliers varied considerably across individuals, with larger tAIs being indicative of more severe symptoms across DSM-5 domains. However, being a neuroanatomical outlier based on tAIs was not specific to ASD. For instance, 84% of the control subjects with mild intellectual disability fell outside the neurotypical range, and tAIs are not only correlated with the genome-wide polygenic risk for ASD (16) but also those for ADHD (28), depression (30), and schizophrenia (29)—phenotypes that are also genetically correlated with ASD (16). Thus, while our composite measure of neuroanatomical atypicality may be highly sensitive to deviations from the neurotypical range, the detected outliers are not specific to diagnostic labels and may detect other conditions associated with atypical neurodevelopment. In a next step, we therefore also examined whether the patterns of cortical thickness differences we observed are linked to ASD etiology.
Similar to the findings of Romero-Garcia et al. (7), we found the t-map of statistical between-group differences to be enriched for genes and coexpression modules that are downregulated in the ASD cortex, which in turn code for synaptic and neuronal proteins (8). Some of these modules (e.g., M12 [34]) show a significant overrepresentation of known ASD-associated genes, which are typically expressed during early (i.e., prenatal) brain development and regulate gene expression at various developmental stages (15, 46). In contrast, the chi-square map of neuroanatomical outliers (and the Cohen’s f effect size map) was enriched predominantly for genes upregulated in ASD, which map onto immune/inflammatory pathways (34) that are most active in the first decades of life (8). Set-based PGSs across sets that are differentially expressed in ASD, and during typical brain development, also explained a larger proportion of tAIs variability than PGSs computed on ASD risk genes (15, 16). This highlights that the t-map and the chi-square outlier map are intrinsically different: the t-map reflects group-level differences, whereas the chi-square map detects outliers within the neurotypical cortical thickness range and can therefore accommodate highly variable patterns of neuroanatomical deviations across individuals. Our findings thus agree with the notion that the effects of ASD susceptibility genes on the brain are pleiotropic, mediated via gene regulatory mechanisms during childhood and adolescence, which result in highly individualized neuroanatomical patterns or “fingerprints.” The examination of these patterns, rather than group differences, may therefore hold the key to stratifying and subtyping ASD.
To this aim, we further established that differences in neuroanatomical deviations are associated with distinct clinical ASD phenotypes that differed in the severity and pattern of sensory symptoms (20). These differences were primarily observed in brain regions subserving sensorimotor control (47), which has been reported to be dysregulated in ASD (48). In these brain regions, individuals with ASD with moderate and severe sensory symptoms displayed predominantly negative cortical thickness deflections (i.e., >72%) compared with individuals with low sensory symptoms. Individuals with ASD might therefore be stratified into biologically more homogeneous subgroups not only based on their absolute phenotypic difference, but also depending on how and to what extent they deviate from the neurotypical mean. Brain regions that differed the most between subgroups were also enriched for genes expressed in excitatory neurons in the developing cortex (38). Despite our sample size not allowing for direct comparison between genotypes and phenotypes, our findings therefore support the notion that a disrupted excitation-inhibition balance (22, 49) may underpin some autism phenotypes (50) and link sensory symptoms to excitatory neurons in the developing cortex. Here, it is important to note that we examined measures of cortical thickness exclusively. Evidence suggests, however, that different aspects of the cortical architecture have distinct genetic determinants (51), contrasting phylogeny (52), and differing developmental trajectories (43). It will therefore be important to repeat our analyses using different morphometric features (e.g., measures of surface area and/or cortical gyrification) to further characterize the complex etiology and neuropathology of ASD.
There are a number of additional limitations. Most importantly, as we did not perform an unbiased out-of-sample validation, our study is based on in-sample estimates of the neurotypical cortical thickness range rather than population norms. Even though we have examined the robustness of our results across variable models (see the online supplement), it will be important to establish the model’s generalization performance using independent samples not only of individuals with ASD but also neurotypical control subjects. Moreover, we examined neuroanatomical outliers based on the general linear model, which made it possible to link group differences to patterns of neuroanatomical deviations within the same framework. Further work is therefore needed to replicate our findings using alternative “normative modeling” approaches—for example, employing Gaussian process regression (13)—that are not confined to linear relationships exclusively. Last, our gene expression decoding analysis was based on the Allen Human Brain Atlas, which is the most comprehensive gene expression atlas to date. However, the Allen atlas is based exclusively on adult donors and provides a coverage that is significantly lower than the spatial resolution of our neuroimaging data. We therefore acknowledge the importance of repeating the analyses in high-resolution age-specific gene-expression data sets, once these become available, to corroborate the important link between molecular and macroscopic pathology in ASD.
1 : Diagnostic heterogeneity in psychiatry: towards an empirical solution. BMC Med 2013; 11:201Crossref, Medline, Google Scholar
2
3 : Neuroimaging in autism spectrum disorder: brain structure and function across the lifespan. Lancet Neurol 2015; 14:1121–1134Crossref, Medline, Google Scholar
4 : Neuroanatomy of autism. Trends Neurosci 2008; 31:137–145Crossref, Medline, Google Scholar
5 : Large-scale analyses of the relationship between sex, age, and intelligence quotient heterogeneity and cortical morphometry in autism spectrum disorder. Mol Psychiatry 2020; 25:614–628Crossref, Medline, Google Scholar
6 : Dissecting the heterogeneous cortical anatomy of autism spectrum disorder using normative models. Biol Psychiatry Cogn Neurosci Neuroimaging 2019; 4:567–578Crossref, Medline, Google Scholar
7 : Synaptic and transcriptionally downregulated genes are associated with cortical thickness differences in autism. Mol Psychiatry 2019; 24:1053–1064Crossref, Medline, Google Scholar
8 : Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 2016; 540:423–427Crossref, Medline, Google Scholar
9 : The EU-AIMS Longitudinal European Autism Project (LEAP): clinical characterisation. Mol Autism 2017; 8:27Crossref, Medline, Google Scholar
10 : Developmental trajectories as autism phenotypes. Am J Med Genet C Semin Med Genet 2015; 169:198–208Crossref, Medline, Google Scholar
11 : The EU-AIMS Longitudinal European Autism Project (LEAP): design and methodologies to identify and validate stratification biomarkers for autism spectrum disorders. Mol Autism 2017; 8:24Crossref, Medline, Google Scholar
12 : A normative modelling approach reveals age-atypical cortical thickness in a subgroup of males with autism spectrum disorder. Commun Biol 2020; 3:486Crossref, Medline, Google Scholar
13 : Fractionating autism based on neuroanatomical normative modeling. Transl Psychiatry 2020; 10:384Crossref, Medline, Google Scholar
14 : An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 2012; 489:391–399Crossref, Medline, Google Scholar
15 : Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 2020; 180:568–584.e23Crossref, Medline, Google Scholar
16 : Identification of common genetic risk variants for autism spectrum disorder. Nat Genet 2019; 51:431–444Crossref, Medline, Google Scholar
17 : Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 2018; 362:eaat8127Crossref, Medline, Google Scholar
18 : Single-cell genomics identifies cell type-specific molecular changes in autism. Science 2019; 364:685–689Crossref, Medline, Google Scholar
19 : Virtual histology of multi-modal magnetic resonance imaging of cerebral cortex in young men. Neuroimage 2020; 218:116968Crossref, Medline, Google Scholar
20 : Dissecting the phenotypic heterogeneity in sensory features in autism spectrum disorder: a factor mixture modelling approach. Mol Autism 2020; 11:67Crossref, Medline, Google Scholar
21 : Heterogeneity of sensory features in autism spectrum disorder: challenges and perspectives for future research. Autism Res 2017; 10:703–710Crossref, Medline, Google Scholar
22 : Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav 2003; 2:255–267Crossref, Medline, Google Scholar
23 : Reduced GABA and altered somatosensory function in children with autism spectrum disorder. Autism Res 2017; 10:608–619Crossref, Medline, Google Scholar
24 : Cortical surface-based analysis, I: segmentation and surface reconstruction. Neuroimage 1999; 9:179–194Crossref, Medline, Google Scholar
25 : Cortical surface-based analysis, II: inflation, flattening, and a surface-based coordinate system. Neuroimage 1999; 9:195–207Crossref, Medline, Google Scholar
26 : Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci USA 2000; 97:11050–11055Crossref, Medline, Google Scholar
27 : A unified statistical approach for determining significant signals in images of cerebral activation. Hum Brain Mapp 1996; 4:58–73Crossref, Medline, Google Scholar
28 : Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet 2019; 51:63–75Crossref, Medline, Google Scholar
29 : Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511:421–427Crossref, Medline, Google Scholar
30 : Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci 2019; 22:343–352Crossref, Medline, Google Scholar
31
32 : Tutorial: a guide to performing polygenic risk score analyses. Nat Protoc 2020; 15:2759–2772Crossref, Medline, Google Scholar
33 : Tight fitting genes: finding relations between statistical maps and gene expression patterns (poster). F1000 Posters, October 28, 2014 (https://f1000research.com/posters/1097120)Google Scholar
34 : Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 2011; 474:380–384Crossref, Medline, Google Scholar
35 : Spatio-temporal transcriptome of the human brain. Nature 2011; 478:483–489Crossref, Medline, Google Scholar
36 : Mapping the genetics of neuropsychological traits to the molecular network of the human brain using a data integrative approach. Biorxiv 2018; 336776Google Scholar
37 : Patterns of sensory processing in children with an autism spectrum disorder. Res Autism Spectr Disord 2014; 8:1214–1224Crossref, Google Scholar
38 : A single-cell transcriptomic atlas of human neocortical development during mid-gestation. Neuron 2019; 103:785–801.e8Crossref, Medline, Google Scholar
39 : Neuroanatomical differences in brain areas implicated in perceptual and other core features of autism revealed by cortical thickness analysis and voxel-based morphometry. Hum Brain Mapp 2010; 31:556–566Medline, Google Scholar
40 : Anatomical differences in the mirror neuron system and social cognition network in autism. Cereb Cortex 2006; 16:1276–1282Crossref, Medline, Google Scholar
41 : Brain surface anatomy in adults with autism: the relationship between surface area, cortical thickness, and autistic symptoms. JAMA Psychiatry 2013; 70:59–70Crossref, Medline, Google Scholar
42 : Age-related temporal and parietal cortical thinning in autism spectrum disorders. Brain 2010; 133:3745–3754Crossref, Medline, Google Scholar
43 : The effect of age, diagnosis, and their interaction on vertex-based measures of cortical thickness and surface area in autism spectrum disorder. J Neural Transm (Vienna) 2014; 121:1157–1170Crossref, Medline, Google Scholar
44 : Cortical thickness abnormalities in autism spectrum disorders through late childhood, adolescence, and adulthood: a large-scale MRI study. Cereb Cortex 2017; 27:1721–1731Crossref, Medline, Google Scholar
45 : Toward a better understanding of neuronal migration deficits in autism spectrum disorders. Front Cell Dev Biol 2019; 7:205Crossref, Medline, Google Scholar
46 : Prenatal origins of ASD: the when, what, and how of ASD development. Trends Neurosci 2020; 43:326–342Crossref, Medline, Google Scholar
47 : Sensorimotor dysfunctions as primary features of autism spectrum disorders. Sci China Life Sci 2015; 58:1016–1023Crossref, Medline, Google Scholar
48 : The role of sensorimotor difficulties in autism spectrum conditions. Front Neurol 2016; 7:124Crossref, Medline, Google Scholar
49 : Excitation-inhibition balance as a framework for investigating mechanisms in neuropsychiatric disorders. Mol Psychiatry 2019; 24:1248–1257Crossref, Medline, Google Scholar
50 : Excitation-inhibition dysbalance as predictor of autistic phenotypes. J Psychiatr Res 2018; 104:96–99Crossref, Medline, Google Scholar
51 : Distinct genetic influences on cortical surface area and cortical thickness. Cereb Cortex 2009; 19:2728–2735Crossref, Medline, Google Scholar
52 : Defects of neuronal migration and the pathogenesis of cortical malformations. Prog Brain Res 1988; 73:15–37Crossref, Medline, Google Scholar



