Genome-Wide Linkage Scan for Loci Predisposing to Social Phobia: Evidence for a Chromosome 16 Risk Locus
Abstract
OBJECTIVE: Social phobia is a common, sometimes disabling, fear of situations that might entail scrutiny by others. Several anxiety disorders, including social phobia, are genetically influenced. Genetic linkage analysis can provide the means to identify genomic locations harboring susceptibility loci for genetically influenced disorders. Identifying loci for social phobia was the goal of this study. METHOD: The authors conducted a genome-wide linkage scan, i.e., tested enough genetic markers to query the entire genome, in 17 American pedigrees (163 subjects) ascertained through probands with panic disorder. Several anxiety disorders segregate in these families; diagnoses were based on structured interviews. A total of 422 markers (404 autosomal, 18 on the X chromosome) with an average spacing of less than 10 centimorgans were genotyped. Multipoint lod score and nonparametric (Zlr score) linkage analyses for social phobia were completed with Allegro and Genehunter X software. RESULTS: Evidence for linkage to social phobia for chromosome 16 markers was identified. A Zlr score of 3.41 was observed for chromosome 16 near marker D16S415. The maximum observed lod score was 2.22, also for chromosome 16, between D16S415 and D16S503 (under a model of recessive inheritance). Additional areas of interest were identified on chromosomes 9, 14, and 18. CONCLUSIONS: These findings meet conservative criteria for “suggestive” linkage. The gene encoding the norepinephrine transporter protein (SLC6A2) maps to this broad region, making SLC6A2 both a positional and physiological candidate for influencing social phobia risk. To the authors’ knowledge, this is the first complete linkage genome scan for this disorder.
Social phobia is a common anxiety disorder (lifetime prevalence =13.3%) (1) that can greatly interfere with normal social and occupational functioning (2–4). According to DSM-III-R diagnostic criteria, social phobia is characterized principally by persistent fear of situations where the individual might be exposed to the scrutiny of others, resulting in either avoidance of such situations or endurance of them with intense anxiety, in a way that interferes with functioning or causes much distress. Like most anxiety disorders, social phobia shows substantial female predominance (3, 5).
Social phobia aggregates in families and is genetically influenced. In studies of adult probands, social phobia has consistently been found to aggregate in their families (6–8), including their children (9). Moreover, a number of social anxiety traits are enriched among first-degree relatives of probands with social phobia (10). There is some evidence that the more severe, generalized subtype of the disorder (i.e., generalized social phobia) shows the strongest tendency toward familial aggregation (6, 8). An alternative explanation for this finding, however, is that generalized social phobia is merely a more severe form of social phobia (11) and that ascertaining for this more severe form of the disorder results in the appearance of greater familial aggregation. One study (12) suggests that social phobia aggregates independently of other phobic disorders (i.e., agoraphobia or simple phobia), although other reports do not support this finding (discussed in the following). A meta-analysis (13) provided a summary odds ratio for the familiality of phobic disorders, including social phobia, of 4.1 (95% confidence interval=2.7–6.1). Taken together, these findings indicate that social phobia has a familial (although not necessarily solely genetic) basis.
Twin studies suggest that a modest portion of the familial resemblance in social anxiety is, indeed, heritable (14–18). Heritability data were reported by Kendler et al. (17), who considered heritability for social phobia, among other anxiety disorders, in a population-based study of female twins with two assessments 8 years apart. Heritability of social phobia was estimated as 51% (corrected for unreliability). A similar heritability estimate (48%) was obtained for a measure of social-anxiety-related cognitions (i.e., fear of negative evaluation) in volunteer twins (19). Kendler et al. (18) reported heritability of 24% for social phobia in a set of male twins (without correction for unreliability). Together, these studies support a moderate role for genetic influences on susceptibility to social phobia (13).
Anxiety disorders co-occur very frequently. In the Epidemiological Catchment Area study, the odds ratios for the diagnosis of social phobia with other anxiety disorders were 3.2 for panic disorder, 11.8 for agoraphobia, and 9.2 for simple phobia (3). In the National Comorbidity Survey, Magee et al. (5) reported that subjects with social phobia had a 23.3% rate of agoraphobia, a 37.6% rate of simple phobia, and a 10.9% rate of panic disorder. Presumably, social phobia shares genetic complexity in common with other psychiatric disorders, perhaps especially other anxiety disorders.
In earlier work, investigators were able to provide evidence against linkage of generalized social phobia to the genes for the serotonin transporter protein and serotonin 2A receptor (20) and a series of dopamine receptor genes (21) in a set of Canadian families. These studies had power only to detect major susceptibility loci, a situation we now know to be unrealistic for what is certain to be a complex trait. Gratacòs et al. (22) identified a chromosome 15 interstitial duplication, which they designated DUP25, that they found to be linked and associated with a range of anxiety disorders (including social phobia), together with joint laxity. They also found evidence for non-Mendelian inheritance. The linkage described was observed between the DUP25 phenotype and various diagnostic constructs, i.e., it was a phenotype-phenotype linkage rather than a marker-phenotype or marker-marker linkage, as is more typically reported. To our knowledge, this finding has not yet been replicated, and its place in our understanding of the genetics of anxiety disorders is potentially great but still uncertain.
We collected a set of families suitable for linkage analysis that would address our primary goal of identifying loci influencing the development of panic disorder. The families segregate not only panic disorder and agoraphobia, but several other anxiety disorders, including social phobia. In previous reports, we considered linkage to panic disorder and agoraphobia (23) and simple phobia (24); in the former article we presented evidence supporting genetic risk factors that were largely specific for panic disorder or agoraphobia, and in the latter we presented strong evidence for linkage of chromosome 14 markers to simple phobia. In this report, we present results for social phobia. We provide herein additional evidence for genetic loci specifically affecting risk for different anxiety disorders, as well as for loci that affect risk for more than one disorder. We believe that this is the first genome-wide linkage scan for social phobia.
Method
Pedigree Identification and Collection of DNA
Most families were first identified through the Anxiety Clinic at the Connecticut Mental Health Center at Yale University. All families in this series were ascertained through probands with panic disorder. A history of two or more additional family members with symptoms of an anxiety disorder, one with panic disorder, was required. Family history inclusion criteria were known symptoms of panic attacks, agoraphobia, generalized anxiety, or social phobia with avoidance. Exclusion criteria included known bilineal affection with anxiety disorders in the parents of probands, certain comorbid diagnoses in probands (history of psychosis, primary alcoholism, or primary substance abuse), insufficient numbers of relatives available for assessment, refusal of the proband to allow relatives to be contacted, or refusal of relatives to participate. All subjects were European-American. For some subjects, immortalized cell lines were created and DNA was extracted from the cell lines; for the remainder, DNA was extracted directly from whole blood. All subjects gave informed consent as approved by the relevant institutional review boards. The procedures for social phobia, described here, are identical to those for panic disorder and agoraphobia (23), as these analyses were carried out in an overlapping set of pedigrees; however, the present series has been augmented and contains genotyped individuals who were not included in our previous studies of panic disorder, agoraphobia, and simple phobia, and it excludes three families with zero linkage information for social phobia.
Diagnosis
The Schedule for Affective Disorders and Schizophrenia—Lifetime Version (SADS-L) (25) (modified version to collect information necessary for DSM-III-R diagnosis) was used in diagnosis for most subjects, but a small number of recently assessed subjects received the Structured Clinical Interview for DSM-IV Axis I Disorders Research Version, Non-Patient Edition (26) (expanded to collect additional anxiety information consistent with what was collected in the modified SADS), instead. Direct interviews were administered to each available individual in an extended pedigree. Family history information on first- and second-degree relatives was obtained from each person interviewed. All proband interviews were conducted face to face. Some interviews for distant subjects were completed by telephone. We used the best-estimate procedure for diagnosis. Research diagnoses were made blindly by one or more psychiatrists (including J.G.) according to DSM-III-R criteria, based on information in the structured interview, notes taken by the interviewer, information about each individual obtained from relatives, and available medical records.
Among the 163 individuals (in 17 pedigrees) considered here, the number of those affected with social phobia was 56 with definite cases and four with probable cases. Power analysis using SIMLINK (27) with 100 replicates showed average Elod scores for social phobia, under the assumption of genetic homogeneity, of 7.1 for a model of recessive inheritance and 8.7 for a model of dominant inheritance.
Markers
Genotypes from 422 markers (18 X chromosome markers, 404 autosomal markers) were analyzed. The autosomal markers included 378 from the Applied Biosystems PRISM LD-MD10 Linkage Mapping Set, version 2.0 (Foster City, Calif.), plus six markers used to replace failed markers from the Applied Biosystems set. We also genotyped polymorphisms (mostly single-nucleotide polymorphisms) at several candidate loci. Finally, additional markers (mostly drawn from the Applied Biosystems PRISM LM-HD5 marker set) were added to increase marker density, either in regions of interest for simple phobia based on an analysis using the 10-cM marker set (chromosome 14 markers) or in regions of interest for panic disorder or agoraphobia based on previous results for those phenotypes. For markers that were on the Marshfield genetic map (28), the sex-averaged distances were used. For the few markers not on the Marshfield map (mostly the candidate polymorphisms), the nucleotide positions of the markers were located in the National Center for Biotechnology Information map viewer (http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi?), and the flanking markers with defined positions on the Marshfield map and map viewer were identified. The genetic distance was then estimated by using the approximation that 1 megabase=1 centimorgan (1 Mb=1 cM). Polymerase chain reaction, genotyping, and allele calling are described elsewhere (23).
Analysis
As described previously (23), the markers were checked for Mendelian segregation errors by using Pedcheck (29) and then PREST (30) to resolve pedigree discrepancies. Separate analyses were done for “definitely” affected subjects (“narrow” definition) and for an expanded set of cases including “probably” affected subjects (“broad” definition). DSM-III-R criteria were used as the basis for diagnosis under both the broad and narrow definitions; the difference was the reviewer’s degree of certainty of that diagnosis.
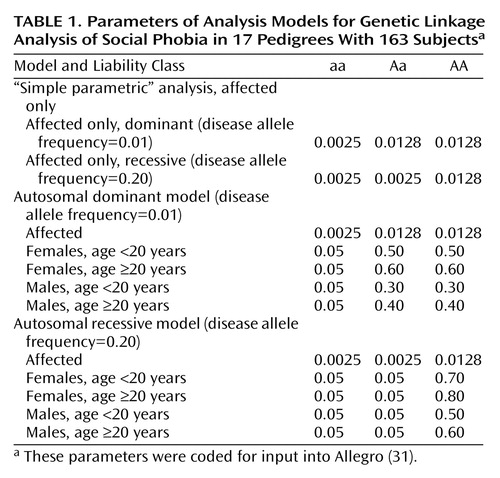
For linkage analysis, we used several stages: fully nonparametric analysis using the Zlr score, “simple parametric” (affected only) analysis (described in more detail in the following), and fully parametric (full model) analysis. Nonparametric Zlr and semiparametric affected-only analyses (Table 1) were completed at two different frequencies of the disease allele (p=0.01 and p=0.20, corresponding to models for dominant and recessive inheritance), and parametric analyses were completed under models of dominant and recessive inheritance (Table 1). In the parametric models, unaffected individuals were modeled with several liability classes based upon age and sex, and values for individuals without data were set to “missing.” The liability classes were derived from published prevalence data (8) and from age-at-onset observations in this series and a previous panic disorder linkage study (32). The models used are consistent with a higher incidence in females than in males, with the majority of incident cases occurring before age 20. Age at onset in the present series was similar to what has been reported previously for this phenotype. Specifically, for the 56 definite cases, there were two onsets between (inclusive) ages 5 and 9 years, 22 between 10 and 14 years, 22 between 15 and 19 years, seven between 20 and 24 years, two between 25 and 29 years, and one between 40 and 44 years.
Allegro (31) was used for all analyses of autosomal markers. Zlr scores are reported rather than NPL scores (for essentially model-free results based on allele sharing) because it has been demonstrated that the NPL scores calculated by Genehunter are too conservative when inheritance data are incomplete, and this problem is addressed by the Genehunter-Plus Zlr algorithm (33) also incorporated in Allegro. Allegro is otherwise similar to Genehunter, but it is faster and allows analysis of somewhat larger families than Genehunter. Lod scores (logarithm of the odds ratio for linkage) and Zlr scores were calculated at each marker and at one point equidistant between each marker pair. Genehunter X (34) was used for X chromosome analyses.
An affected-only analysis involves setting penetrances low and disease allele frequencies high and analyzing only the phenotypes for affected subjects, with the others set to “unknown phenotype.” These choices reduce some of the difficulties with lod score analysis when the genetic model is not exactly known, because assumptions about unaffected individuals can be reduced (35). (In general, applying model-based linkage analysis when the model is unknown will primarily result in false negative rather than false positive results [36].) The model used in these analyses is based on the assumption that the locus being mapped accounts for only a fraction of the population-level risk for the phenotype and that there are multiple phenocopies and sporadic cases.
Results
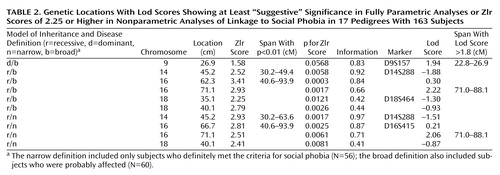
Linkage results for social phobia for the fully parametric model (lod score) and nonparametric model (Zlr score) (Table 2) are summarized in Figure 1 and Figure 2. Two regions provided statistical support for linkage at the “suggestive” level (37). The first of these, on chromosome 16, covers an extensive region. The maximum lod score observed in this region was 2.22 (recessive/broad model) at position 71.1. This was also the highest lod score observed in the present study. The recessive/narrow model gave a lod score of 2.06 at the same location. The maximum Zlr score observed in this region, and the highest for this study, was 3.41, at position 62.3 (p=0.0003). This Zlr score is included in a region from position 40.6 to 93.9 where the Zlr score is greater than 1, a region of 53.3 cM (Figure 3). The second, on chromosome 9 at marker D9S157, showed a lod score of 1.94 (dominant/broad model). The corresponding Zlr score was 1.58.
Other regions of interest were identified on chromosomes 14 and 18. On chromosome 14, Zlr scores of 2.93 and 2.52 were observed at marker D14S288 (position 45.2) under narrow and broad disease definitions, respectively. However, the lod score at this position was negative. On chromosome 18, Zlr scores of 2.79 (p=0.0026) and 2.41 (p=0.0081) were observed at position 40.1, under broad and narrow disease definitions, respectively.
In the affected-only (or “simple parametric”) analysis (Table 3), heterogeneity lod (Hlod) scores above 2 were observed for locations on chromosomes 14 and 16. On chromosome 14, Hlod scores of 2.82 (α=0.21) and 2.06 (α=0.36) were observed at markers D14S276 and D14S288, respectively, under the p=0.01/narrow and p=0.01/broad models, respectively. A lod score of 2.47 was observed for the p=0.01/narrow model as well at position 43.3, close to D14S276. On chromosome 16, an Hlod score of 2.66 (α=0.22) and a lod score of 2.48 were observed at position 51.8 (marker D16S3136). No lod or Hlod score above 1.8 was observed under the p=0.20 affected-only model.
The strongest evidence for linkage was thus found on chromosome 16, with lod score 2.48 and Hlod score 2.66 (simple parametric analysis, dominant/narrow), lod score 2.22 (recessive/broad), and lod score 2.06 (recessive/narrow), together with Zlr score 3.41, all observed within a span of about 20 cM. These results taken together may be considered strongly suggestive of linkage to this region.
Discussion
To our knowledge, this is the first genome-wide scan linkage study of social phobia. The strongest evidence for linkage to social phobia was on chromosome 16. A Zlr score of 3.41 was observed for chromosome 16 near marker D16S415 (p=0.0003), in the context of lod scores higher than 2 under several models (with the most notable parametric model results based on the assumption of recessive inheritance). The most obvious candidate gene mapped in this broad region is SLC6A2 (“solute carrier family 6 member 2”), the norepinephrine transporter protein locus (protein product, NET1), which maps close to D16S3136.
The norepinephrine transporter plays the physiological role of reuptake of norepinephrine and dopamine into presynaptic neurons, terminating neurotransmission. It is a target for some medications that are effective for the treatment of social anxiety disorder, including venlafaxine (38) and, possibly, paroxetine (39). Adrenergic tone clearly plays a role in the experience of some of the physiological concomitants of anxiety, and blockade of norepinephrine can be involved in relief of some symptoms of anxiety. Furthermore, there is preliminary evidence that dopamine metabolism may be abnormal in social phobia (40). These observations make SLC6A2 (which succeeded “SLC6A5” as the locus symbol for this gene) a clear candidate gene for anxiety disorders in general (41) and social phobia in particular.
We are aware of two previous linkage studies of social phobia (20, 21). Those reports considered markers at a total of six candidate loci in a group of subjects somewhat smaller than the present study group, without evidence of linkage; SLC6A2 was not among the candidate genes evaluated.
We may consider all of the linkage results presented herein in the context of results for other anxiety disorders, sometimes comorbid with social phobia and with each other, obtained in the larger group of subjects from which the present study group was drawn. Chromosome 16 markers generated Hlod scores in the range of 1.1–1.4 and Zlr scores as high as 2.51 for the simple phobia phenotype (24) but no lod, Hlod, or nonparametric linkage scores of interest (NPL scores, in this case) for panic disorder or agoraphobia (23).
The region of interest for social phobia on chromosome 9 does not overlap with regions of interest for simple phobia, panic disorder, or agoraphobia. Hlod scores for this region were high consistently across all parametric models (Figure 2). The region of interest for social phobia on chromosome 14 (Hlod score=2.82 under the dominant/narrow model, Zlr score=2.52 under a broad model) (Figure 1 and Figure 2) overlaps with a region of significant linkage for simple phobia we reported earlier (lod score as high as 3.70 under a simple parametric model) (24). This same region also generated NPL scores higher than 2 for both panic disorder and agoraphobia (23). The region of interest for social phobia on chromosome 18 (Zlr score=2.41 under a model with a narrow diagnosis definition) does not find any corresponding positive linkage values for panic disorder or agoraphobia, but we previously (24) reported a Zlr score of 1.84 for simple phobia (model with narrow diagnosis definition) about 10 cM distant.
The regions of greatest interest for panic disorder (chromosomes 1 and 11) and agoraphobia (chromosome 3) (23) were not identified as areas of interest for social phobia. Thus, the results, considered together, suggest that a chromosome 16 locus may be important in determining social phobia risk, is less likely to be important for simple phobia risk, and does not contribute to panic disorder or agoraphobia. Conversely, the loci that were most likely to be important for the risk for panic disorder and agoraphobia do not appear to be important for social phobia. Finally, a chromosome 14 locus that is significantly linked to simple phobia may be of lesser importance for social phobia, panic disorder, and agoraphobia. Of all of the possible linkages identified in parts of this group of subjects, the putative chromosome 14 locus is most likely to play a role in susceptibility to a range of anxiety-related phenotypes. These observations are consistent with a hypothesis suggested by patterns of comorbidity among the anxiety disorders and presented by us (23) and earlier by Kendler et al. (42) in a broader context, that there may be risk loci that increase risk for several anxiety disorders and other risk loci that are more specific to certain anxiety disorders. Without the existence of such shared risk loci, it is difficult to explain the elevated rates of co-segregating social and simple phobia in pedigrees recruited not for these disorders but for high rates of panic disorder.
It is possible that social phobia as observed in this set of families selected for panic disorder may represent a genetic subtype that is different from social phobia as observed in the general population or in families selected specifically for aggregation of social phobia. If this is the case, the findings may not generalize to a differently ascertained study group. This strategy may also have decreased genetic heterogeneity for social phobia in our particular group, and such a decrease would be expected to be advantageous for gene mapping for those specific loci enriched in the group.
Power analysis suggested that this study group had adequate power to detect linkage when the phenotype studied was inherited in a simple Mendelian fashion, which was clearly a liberal assumption. Not surprisingly, our data support complex inheritance for this common disorder. The size of the group was only small to moderate for mapping a complex trait, but the power is made greater than that for a disorder with later onset (such as panic disorder) by the relatively greater informativeness of younger subjects due to a more favorable age-corrected liability curve for social phobia. Also, an extended-pedigree design, such as we employed, provides greater power for a given group size than many other designs used to study the genetics of complex traits, such as affected sibling pair designs.
A confluence of strong “suggestive” results under different analytic models, taken together with the presence in the genomic region of a favored candidate gene (SLC6A2, which encodes NET1) (41), supports the possibility of a social phobia risk locus on chromosome 16. These results require confirmation in additional groups of subjects. Ideally, this would occur in subjects collected through social phobia probands. A future study could also be enhanced by consideration of quantitative or presumptive intermediate phenotypes, such as heart rate, plasma norepinephrine level or other catecholamine index (such as 3-methoxy-4-hydroxyphenylglycol), or imaging measures that offer an opportunity to use specific ligands to directly image the norepinephrine transporter.
 |
 |
 |
Received Sept. 13, 2002; revision received Jan. 16, 2003; accepted March 2, 2003. From the Department of Psychiatry, Yale University School of Medicine, New Haven, Conn.; the Department of Psychiatry, VA Connecticut Healthcare Center, West Haven; the Department of Biometry and Epidemiology, Medical University of South Carolina, Charleston; and the Department of Psychiatry, University of California, San Diego, School of Medicine. Address reprint requests to Dr. Gelernter, Yale University School of Medicine, VA Psychiatry 116A2; 950 Campbell Ave., West Haven, CT 06516; [email protected] (e-mail). Supported in part by the U.S. Department of Veterans Affairs through the VA Medical Research Program (Merit Review to Dr. Gelernter), the VA Connecticut-Massachusetts Mental Illness Research, Education, and Clinical Center, and the VA Connecticut Research Enhancement Award Program; by grant MH-01387 from NIMH; and by grants DA-12690 and DA-12849 from the National Institute on Drug Abuse. The authors thank Kathleen Bonvicini, M.P.H., for interviewing assistance; Greg Kay and Anne Marie Lacobelle for technical assistance; Susan Kruger, M.D., and Susan Goodson, M.D., for assistance with diagnoses; and the laboratory of Dr. K. Kidd for establishment of cell lines for many of the samples studied.
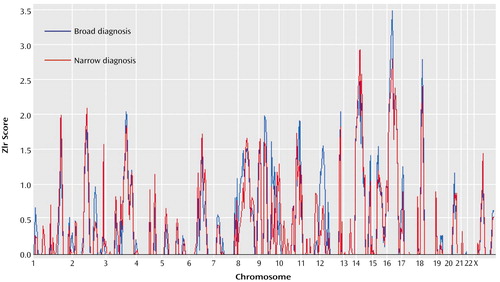
Figure 1. Genome-Wide Results of Nonparametric Analyses of Genetic Linkage of Social Phobia in 17 Pedigrees With 163 Subjects, Based on Broad and Narrow Definitions of Social Phobiaa
aThe narrow definition included only subjects who definitely met the criteria for social phobia (N=56); the broad definition also included subjects who were probably affected (N=60).
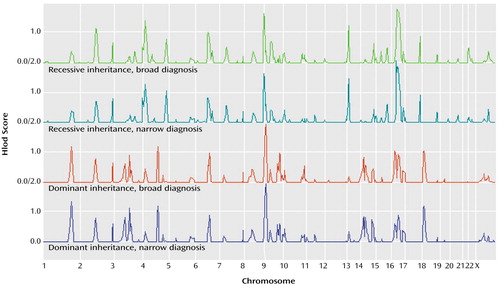
Figure 2. Genome-Wide Heterogeneity Lod (Hlod) Scores for Four Parametric Models of Genetic Linkage of Social Phobia in 17 Pedigrees With 163 Subjects, Based on Dominant and Recessive Inheritance and Broad and Narrow Definitions of Social Phobiaa
aThe narrow definition included only subjects who definitely met the criteria for social phobia (N=56); the broad definition also included subjects who were probably affected (N=60).
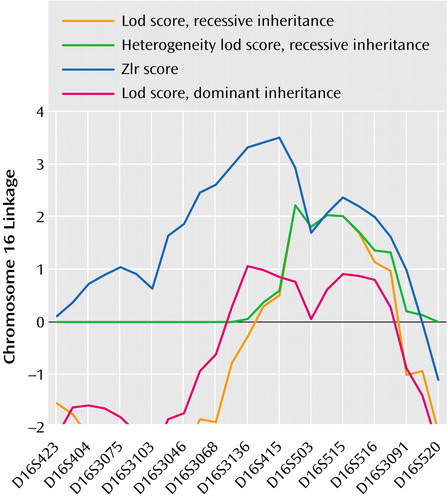
Figure 3. Results of Nonparametric Analyses (Zlr scores) and Parametric Analyses (lod scores) of Linkage of Markers on Chromosome 16 to Social Phobia in 17 Pedigrees With 163 Subjects
1. Kessler RC, McGonagle KA, Zhao S, Nelson CB, Hughes M, Eshleman S, Wittchen H-U, Kendler KS: Lifetime and 12-month prevalence of DSM-III-R psychiatric disorders in the United States: results from the National Comorbidity Survey. Arch Gen Psychiatry 1994; 51:8–19Crossref, Medline, Google Scholar
2. Gelernter CS, Uhde TW, Cimbolic P, Arnkoff DB, Vittone BJ, Tancer ME, Bartko JJ: Cognitive-behavioral and pharmacological treatments of social phobia: a controlled study. Arch Gen Psychiatry 1991; 48:938–945Crossref, Medline, Google Scholar
3. Schneier FR, Johnson J, Hornig CD, Liebowitz MR, Weissman MM: Social phobia: comorbidity and morbidity in an epidemiologic sample. Arch Gen Psychiatry 1992; 49:282–288Crossref, Medline, Google Scholar
4. Stein MB, Kean YM: Disability and quality of life in social phobia: epidemiologic findings. Am J Psychiatry 2000; 157:1606–1613Link, Google Scholar
5. Magee W, Eaton WW, Wittchen H-U, McGonagle KA, Kessler RC: Agoraphobia, simple phobia, and social phobia in the National Comorbidity Survey. Arch Gen Psychiatry 1996; 53:159–168Crossref, Medline, Google Scholar
6. Mannuzza S, Schneier FR, Chapman TF, Liebowitz MR, Klein DF, Fyer AJ: Generalized social phobia: reliability and validity. Arch Gen Psychiatry 1995; 52:230–237Crossref, Medline, Google Scholar
7. Nelson EC, Grant JD, Bucholz KK, Glowinski A, Madden PAF, Reich W, Heath AC: Social phobia in a population-based female adolescent twin sample: co-morbidity and associated suicide-related symptoms. Psychol Med 2000; 30:797–804Crossref, Medline, Google Scholar
8. Stein MB, Chartier MJ, Hazen AL, Kozak MV, Tancer ME, Lander S, Furer P, Chubaty D, Walker JR: A direct-interview family study of generalized social phobia. Am J Psychiatry 1998; 155:90–97Link, Google Scholar
9. Mancini C, van Ameringen M, Szatmari P, Fugere C, Boyle MA: A high-risk pilot study of the children of adults with social phobia. J Am Acad Child Adolesc Psychiatry 1996; 35:1511–1517Crossref, Medline, Google Scholar
10. Stein MB, Chartier MJ, Lizak MV, Jang KL: Familial aggregation of anxiety-related quantitative traits in generalized social phobia: clues to understanding “disorder” heritability? Am J Med Genet 2001; 105:79–83Crossref, Medline, Google Scholar
11. Stein MB, Torgrud LJ, Walker JR: Social phobia symptoms, subtypes and severity: findings from a community survey. Arch Gen Psychiatry 2000; 57:1046–1052Crossref, Medline, Google Scholar
12. Fyer AJ, Mannuzza S, Chapman TF, Martin LY, Klein DF: Specificity in familial aggregation of phobic disorders. Arch Gen Psychiatry 1995; 52:564–573Crossref, Medline, Google Scholar
13. Hettema JM, Neale MC, Kendler KS: A review and meta-analysis of the genetic epidemiology of anxiety disorders. Am J Psychiatry 2001; 158:1568–1578Link, Google Scholar
14. Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ: The genetic epidemiology of phobias in women: the interrelationship of agoraphobia, social phobia, situational phobia, and simple phobia. Arch Gen Psychiatry 1992; 49:273–281Crossref, Medline, Google Scholar
15. Skre I, Onstad S, Torgersen S, Lygren S, Kringlen E: A twin study of DSM-III-R anxiety disorders. Acta Psychiatr Scand 1993; 88:85–92Crossref, Medline, Google Scholar
16. Torgersen S: Genetic factors in anxiety disorders. Arch Gen Psychiatry 1983; 40:1085–1089Crossref, Medline, Google Scholar
17. Kendler KS, Karkowski LM, Prescott CA: Fears and phobias: reliability and heritability. Psychol Med 1999; 29:539–553Crossref, Medline, Google Scholar
18. Kendler KS, Myers J, Prescott CA, Neale MC: The genetic epidemiology of irrational fears and phobias in men. Arch Gen Psychiatry 2001; 58:257–265Crossref, Medline, Google Scholar
19. Stein MB, Jang KL, Livesley WJ: Heritability of social anxiety-related concerns and personality characteristics: a twin study. J Nerv Ment Dis 2002; 190:219–224Crossref, Medline, Google Scholar
20. Stein MB, Chartier MJ, Kozak MV, King N, Kennedy JL: Genetic linkage to the serotonin transporter protein and 5HT2A receptor genes excluded in generalized social phobia. Psychiatry Res 1998; 81:283–291Crossref, Medline, Google Scholar
21. Kennedy JL, Neves-Pereira M, King N, Lizak MV, Basile VS, Chartier MJ, Stein MB: Dopamine system genes not linked to social phobia. Psychiatr Genet 2001; 11:213–217Crossref, Medline, Google Scholar
22. Gratacòs M, Nadal M, Martin-Santos R, Pujana MA, Gago J, Peral B, Armengol L, Ponsa I, Miro R, Bulbena A, Estivill X: A polymorphic genomic duplication on human chromosome 15 is a susceptibility factor for panic and phobic disorders. Cell 2001; 106:367–379Crossref, Medline, Google Scholar
23. Gelernter J, Bonvicini K, Page G, Woods SW, Goddard AW, Kruger S, Pauls DL, Goodson S: Linkage genome scan for loci predisposing to panic disorder or agoraphobia. Am J Med Genet Neuropsychiatr Genet 2001; 105:548–557Crossref, Medline, Google Scholar
24. Gelernter J, Page G, Bonvicini K, Woods SW, Pauls DL, Kruger S: A chromosome 14 risk locus for simple phobia: results from a genomewide linkage scan. Mol Psychiatry 2003; 8:71–82Crossref, Medline, Google Scholar
25. Spitzer RL, Endicott J: Schedule for Affective Disorders and Schizophrenia—Lifetime Version. New York, New York State Psychiatric Institute, Biometrics Research, 1975Google Scholar
26. First MB, Spitzer RL, Gibbon M, Williams JBW: Structured Clinical Interview for DSM-IV Axis I Disorders Research Version, Non-Patient Edition (SCID-NP), version 2. New York, New York State Psychiatric Institute, Biometrics Research, 1997Google Scholar
27. Boehnke M: Estimating the power of a proposed linkage study: a practical computer simulation approach. Am J Hum Genet 1986; 39:513–527Medline, Google Scholar
28. Broman KW, Murray JC, Sheffield VC, White RL, Weber JL: Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genetics 1998; 63:861–869Crossref, Medline, Google Scholar
29. O’Connell JR, Weeks DE: PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 1998; 63:259–266Crossref, Medline, Google Scholar
30. McPeek MS, Sun L: Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet 2000; 66:1076–1094Crossref, Medline, Google Scholar
31. Gudbjartsson DF, Jonasson K, Frigge ML, Kong A: Allegro, a new computer program for multipoint linkage analysis (letter). Nat Genet 2000; 25:12–13Crossref, Medline, Google Scholar
32. Knowles JA, Fyer AJ, Vieland VJ, Weissman MM, Hodge SE, Heiman GA, Haghighi F, de Jesus GM, Rassnick H, Preud’homme-Rivelli X, Austin T, Cunjak J, Mick S, Fine LD, Woodley KA, Das K, Maier W, Adams PB, Freimer NB, Klein DF, Gilliam TC: Results of a genome-wide genetic screen for panic disorder. Am J Med Genet Neuropsychiatr Genet 1998; 81:139–147Crossref, Medline, Google Scholar
33. Kong A, Cox NJ: Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 1997; 61:1179–1188Crossref, Medline, Google Scholar
34. Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES: Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 1996; 58:1347–1363Medline, Google Scholar
35. Xu J, Meyers DA, Pericak-Vance MA: Lod score analysis, in Approaches to Gene Mapping in Complex Human Diseases. Edited by Haines JL, Pericak-Vance MA. New York, Wiley-Liss, 1998, pp 253–272Google Scholar
36. Clerget-Darpoux F, Bonaiti-Pellie C, Hochez J: Effects of misspecifying genetic parameters in lod score analysis. Biometrics 1986; 42:393–399Crossref, Medline, Google Scholar
37. Lander E, Kruglyak L: Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nature Gen 1995; 11:241–247Crossref, Medline, Google Scholar
38. Gorman JM, Kent JM: SSRIs and SNRIs: broad spectrum of efficacy beyond major depression. J Clin Psychiatry 2002; 60(suppl 4):33–38Google Scholar
39. Owens MJ, Knight D, Nemeroff CB: Paroxetine binding to the rat norepinephrine transporter in vivo. Biol Psychiatry 2000; 47:842–845Crossref, Medline, Google Scholar
40. Tiihonen J, Kuikka J, Bergström K, Lepola U, Koponen H, Leinonen E: Dopamine reuptake site densities in patients with social phobia. Am J Psychiatry 1997; 154:239–242Link, Google Scholar
41. Gelernter J, Kruger S, Pakstis AJ, Pacholczyk T, Sparkes RS, Kidd KK, Amara S: Assignment of the norepinephrine transporter protein (NET1) locus to chromosome 16. Genomics 1993; 18:690–692Crossref, Medline, Google Scholar
42. Kendler KS, Walters EE, Neale MC, Kessler RC, Heath AC, Eaves LJ: The structure of the genetic and environmental risk factors for six major psychiatric disorders in women: phobia, generalized anxiety disorder, panic disorder, bulimia, major depression, and alcoholism. Arch Gen Psychiatry 1995; 52:374–383Crossref, Medline, Google Scholar



