Genome-Wide Association Study Meta-Analysis of the Alcohol Use Disorders Identification Test (AUDIT) in Two Population-Based Cohorts
Abstract
Objective:
Alcohol use disorders are common conditions that have enormous social and economic consequences. Genome-wide association analyses were performed to identify genetic variants associated with a proxy measure of alcohol consumption and alcohol misuse and to explore the shared genetic basis between these measures and other substance use, psychiatric, and behavioral traits.
Method:
This study used quantitative measures from the Alcohol Use Disorders Identification Test (AUDIT) from two population-based cohorts of European ancestry (UK Biobank [N=121,604] and 23andMe [N=20,328]) and performed a genome-wide association study (GWAS) meta-analysis. Two additional GWAS analyses were performed, a GWAS for AUDIT scores on items 1–3, which focus on consumption (AUDIT-C), and for scores on items 4–10, which focus on the problematic consequences of drinking (AUDIT-P).
Results:
The GWAS meta-analysis of AUDIT total score identified 10 associated risk loci. Novel associations localized to genes including JCAD and SLC39A13; this study also replicated previously identified signals in the genes ADH1B, ADH1C, KLB, and GCKR. The dimensions of AUDIT showed positive genetic correlations with alcohol consumption (rg=0.76–0.92) and DSM-IV alcohol dependence (rg=0.33–0.63). AUDIT-P and AUDIT-C scores showed significantly different patterns of association across a number of traits, including psychiatric disorders. AUDIT-P score was significantly positively genetically correlated with schizophrenia (rg=0.22), major depressive disorder (rg=0.26), and attention deficit hyperactivity disorder (rg=0.23), whereas AUDIT-C score was significantly negatively genetically correlated with major depressive disorder (rg=−0.24) and ADHD (rg=−0.10). This study also used the AUDIT data in the UK Biobank to identify thresholds for dichotomizing AUDIT total score that optimize genetic correlations with DSM-IV alcohol dependence. Coding individuals with AUDIT total scores ≤4 as control subjects and those with scores ≥12 as case subjects produced a significant high genetic correlation with DSM-IV alcohol dependence (rg=0.82) while retaining most subjects.
Conclusions:
AUDIT scores ascertained in population-based cohorts can be used to explore the genetic basis of both alcohol consumption and alcohol use disorders.
Alcohol use disorders are modestly heritable, with twin studies demonstrating that approximately 50% of the phenotypic variance is attributed to genetic factors (1, 2). To date, genetic studies of alcohol use disorders have identified genes that influence pharmacokinetic factors (e.g., ADH1B, ADH1C, ALDH2) (3–8) but none that influence pharmacodynamic factors. The difficulty of assembling large, carefully diagnosed cohorts of individuals with alcohol use disorders has stimulated additional studies of nonclinical phenotypes, such as alcohol consumption, in populations not ascertained for alcohol dependence. This approach has allowed for the relatively rapid collection of much larger sample sizes (e.g., >100,000 individuals) and has identified numerous loci associated with both pharmacokinetic and pharmacodynamic factors that influence alcohol consumption, including ADH1B/ADH1C/ADH5 (9–11), KLB (encoding β-klotho) (9, 11, 12), and GCKR, encoding the glucokinase regulatory protein (9, 11). However, the genetic overlap between alcohol consumption (in units per week) and diagnosed DSM-IV alcohol dependence is moderate (rg=0.38) (13), which reinforces the notion that alcohol consumption cannot be used as a surrogate for alcohol use disorders.
The Alcohol Use Disorders Identification Test (AUDIT) is a screening tool designed to identify past-year hazardous alcohol use (14). The test consists of 10 items across three dimensions—those pertaining to alcohol consumption (items 1–3, often termed AUDIT-C), dependence symptoms (items 4–6), and problematic or hazardous alcohol use (items 7–10). When the AUDIT was developed, a total score ≥8 was proposed to be indicative of hazardous alcohol use (14) and a score ≥20 consistent with a diagnosis of alcohol dependence (15). However, there is no clear consensus on the threshold for alcohol dependence, and subsequent studies have suggested that additional factors, including sex and cultural and social contexts, should be considered when deriving thresholds for alcohol dependence (reviewed in Table S1 in the online supplement).
A recent population-based GWAS of AUDIT scores in 20,328 research participants from the genetics company 23andMe identified a locus near the gene ADH1C (rs141973904; p=4.4×10−7) (10) that was nominally associated with AUDIT total score. AUDIT scores among 23andMe research participants were low and predominantly driven by alcohol consumption (AUDIT-C score). The genetic correlation between AUDIT total score from the 23andMe sample and alcohol consumption was much stronger (rg=0.89, p=9.01×10−10) than the genetic correlation between AUDIT total score and alcohol dependence (rg=0.08; p=0.65) (13).
In this study, we performed a GWAS meta-analysis using the UK Biobank cohort (N=121,604) and the previously published 23andMe cohort (N=20,328) (10), yielding the largest GWAS meta-analysis of AUDIT total score to date (N=141,932). Using only the UK Biobank cohort, we also sought to determine whether the alcohol consumption component of the AUDIT had a genetic architecture distinct from the dependence and hazardous-use components by performing GWASs of consumption (items 1–3, AUDIT-C) and problems (items 4–10, AUDIT-P). Linkage disequilibrium score regression (16) was used to calculate genetic correlations between AUDIT measures and other substance use, psychiatric, and behavioral traits. We also calculated genetic correlations with obesity and blood lipid traits, as these have previously been shown to be associated with alcohol consumption (9, 10). We hypothesized that AUDIT-P score would correlate more strongly with measures of hazardous substance use, including alcohol dependence, and other psychiatric conditions. Finally, to determine the thresholds for dichotomizing AUDIT total score that would most closely approximate alcohol dependence, we used continuous AUDIT total score to categorize participants as case and control subjects using different thresholds, performed GWAS on each, and calculated the genetic correlation with DSM-IV alcohol dependence (13).
Method
UK Biobank Sample
The UK Biobank (www.ukbiobank.ac.uk) is a population-based sample of 502,629 individuals who were recruited from 22 assessment centers across the United Kingdom from 2006 to 2010 (17). A total of 157,366 individuals completed a mental health questionnaire as part of an online follow-up over a 1-year period in 2017. The AUDIT was administered to assess alcohol use over the past year, using gating logic (see Figure S1 in the online supplement). After quality control procedures were performed to remove participants with missing data, and keeping only white British unrelated individuals, 121,604 individuals with AUDIT total scores were available. AUDIT total score was created by taking the sum of items 1–10, for all participants, including those who endorsed currently never drinking alcohol (as they could still endorse past hazardous use on items 9 and 10). We also created AUDIT subdomain scores by aggregating the scores from items 1–3, which include the information pertaining to alcohol consumption (AUDIT-C, N=121,604), and from items 4–10, which index the information pertaining to alcohol problems (AUDIT-P, N=121,604). These traits were log10-transformed to approximate a normal distribution (see Figure S2 in the online supplement).
Genotyping, Quality Control, and Imputation
Genotype imputation was performed on 487,409 individuals by the UK Biobank team, using IMPUTE4 (18) and the Haplotype Reference Consortium reference panel. After quality control, 16,213,998 single-nucleotide polymorphisms (SNPs) remained for GWAS analyses. Additional details on genotyping and quality control are provided in the Supplementary Methods section in the online supplement.
Discovery GWASs Using the UK Biobank Sample
GWAS analyses were performed using BGENIE, version 1.1 (18), with AUDIT scores (total score, AUDIT-C score, and AUDIT-P score, tested independently) as the outcome variable and age, sex, genotyping array, and the first 20 principal components derived from genotype data as covariates. (See the Supplementary Methods section in the online supplement for more details.) To identify independently associated variants (“index variants”), clump-based pruning was applied in FUMA (fuma.ctglab.nl) using an r2 of 0.1 and a 1-Mb sliding window using the UK Biobank white British sample as the linkage disequilibrium (LD) reference panel. A 1-Mb window was used because of the regions of extended LD on chromosomes 4q23 and 17q21.31, which were associated with AUDIT scores in this study.
In addition, we performed a series of 18 case-control GWAS analyses of AUDIT total score using different thresholds (cases: ≥8, 10, 12, 15, 18, 20; compared with controls: ≤2, 3, 4). The sample size at each threshold is listed in Table S2 in the online supplement. The results of these analyses were used to determine which thresholds would produce the highest genetic correlation estimates with DSM-IV alcohol dependence (13).
SNP Heritability Analyses
The SNP heritability of UK Biobank AUDIT scores (total, AUDIT-C, AUDIT-P) was calculated using a genomic restricted maximum likelihood (GREML) method implemented in Genetic Complex Trait Analysis (GCTA) (19) on a subset of 117,072 unrelated individuals, using a relatedness cutoff of 0.05 and controlling for age and sex. GREML analyses were run using genotyped SNPs with a minor allele frequency greater than 0.01 to construct the genetic relationship matrix.
GWAS Meta-Analysis of AUDIT Total Score Using the UK Biobank and 23andMe Samples
Because the genetic correlation of AUDIT total score between the UK Biobank and 23andMe cohorts was high (rg=0.77, SE=0.12, p=7.15×10−11), we performed a sample size–based meta-analysis of AUDIT total score from the UK Biobank and 23andMe cohorts using METAL (version 2011-03-25) (20). This meta-analysis comprises a total of 141,932 research participants of European ancestry and 9,519,872 genetic variants that passed quality control. We used clump-based pruning (see the “Discovery GWASs Using the UK Biobank” section, above) to identify independently associated variants. For each GWAS signal, we defined a set of credible variants using a Bayesian refinement method developed by Maller et al. (21). These credible sets are considered to have a 99% probability of containing the “causal” variant at each locus. Credible set analyses were performed in R (https://github.com/hailianghuang/FM-summary) for each of the index variants associated with AUDIT total score in the GWAS meta-analysis using SNPs within 1 Mb with an r2 >0.4 to the index variant. All downstream genetic analyses of AUDIT total score were performed using the GWAS meta-analysis summary statistics. The 23andMe AUDIT GWAS has been published previously (10), and 30,441 participants from the UK Biobank cohort were included in a previous GWAS of alcohol consumption (9).The GWAS of alcohol consumption in the UK Biobank sample involved individuals genotyped during the first UK Biobank data release (9). The individuals from the UK Biobank in the present study are those who completed the mental health questionnaire in 2017 and include those genotyped in both the first and second UK Biobank data releases; the sample overlap between these two data sets is 30,441 individuals.
Functional Mapping and Annotation of GWAS Meta-Analysis
We used FUMA, version 1.2.8 (22), to study the functional consequences of the index SNPs and of the SNPs contained in each credible set, which included categories from the ANNOVAR tool, combined annotation-dependent depletion (CADD) scores, RegulomeDB scores (www.regulomedb.org/), expression quantitative trait loci, and chromatin states. We also studied the regulatory consequences of the index SNPs using the Variant Effect Predictor tool (VEP; Ensembl GRCh37).
Gene-Set and Pathway Analyses
We performed MAGMA (22) competitive gene-set and pathway analyses using the summary statistics from the GWAS meta-analysis of AUDIT total score and the AUDIT-C and AUDIT-P score subsets. SNPs were mapped to 18,546 protein-coding genes from Ensembl, build 85. Gene sets were obtained from Msigdb, version 5.2 (“Curated gene sets,” “GO terms”).
Gene-Based Association Using Transcriptomic Data With S-PrediXcan
We used S-PrediXcan (23) to predict gene expression levels in 10 brain tissues and to test whether the predicted gene expression correlates with AUDIT scores. We used precomputed tissue weights from the Genotype-Tissue Expression (version 7) project database (https://www.gtexportal.org/) as the reference transcriptome data set. Further details are provided in the Supplementary Methods section in the online supplement.
Genetic Correlation Analysis
We used LD score regression (LDSC) (github.com/bulik/ldsc; ldsc.broadinstitute.org) to identify genetic correlations between traits (24). This method was used to calculate genetic correlations (rg) between AUDIT total score, AUDIT-C score, and AUDIT-P score, and 39 other traits and diseases (see Tables S3, S4, and S5 in the online supplement). We did not constrain the intercepts in our analysis, as we could not quantify the exact amount of sample overlap between cohorts. We used false discovery rate to correct for multiple testing (25). We also used LDSC to examine genetic correlations between various dichotomized versions of AUDIT and DSM-IV alcohol dependence (13). To test for significant differences between the genetic correlations, z-score statistics were calculated (see Table S6 in the online supplement).
Results
UK Biobank Sample Characteristics
In the UK Biobank cohort, there were 121,604 individuals with AUDIT scores available for GWAS analysis (see Table S7 in the online supplement). The UK Biobank sample was 56.2% female (N=68,389), and the mean age was 56.1 years (SD=7.7). The mean AUDIT total score was 5.0 (SD=4.18, range=0–40); a histogram showing the distribution of the scores is provided in Figure S2 in the online supplement. Over the previous year, 91.9% of the participants reported drinking 1 or 2 drinks on a single drinking day. Over the previous year, 6.3% of the participants reported that they were not able to stop drinking once they started, and 10.7% felt guilt or remorse after drinking (see Table S7 in the online supplement). The mean AUDIT total score was significantly higher for males than for females (6.09 [SD=4.45] and 4.15 [SD=3.72], respectively; β=0.47, p<2×10−6) (see Figure S3 in the online supplement). In addition, age was negatively correlated with AUDIT scores (β=−0.02, p<2×10−6) (see Table S8 in the online supplement). Therefore, both sex and age were used as covariates in the GWAS analyses. The mean AUDIT-C score was 4.24 (SD=2.83) and the mean AUDIT-P score was 0.75 (SD=2.0). As expected, there was a moderate positive phenotypic correlation between AUDIT-C and AUDIT-P scores (r=0.478, 95% CI=0.473–0.481, p<2×10−16) (see Table S8 in the online supplement).
SNP Heritability in the UK Biobank Sample
We estimated the SNP heritability of AUDIT total score to be 12% (GCTA: SE=0.48%, p=4.6×10−273; LDSC: 8.6%, SE=0.50%), which is similar to the previously published AUDIT estimate (10). The SNP heritability for AUDIT-C score was 11% (GCTA: SE=0.47%, p=1.5×10−211; LDSC: 8.4%, SE=0.55%), and 9% for AUDIT-P score (GCTA: SE=0.46%, p=2.0×10−178; LDSC: 5.9%, SE=0.48%).
GWAS of AUDIT Scores in the UK Biobank Sample
The significant results (p<5×10−8) of the GWAS of AUDIT total score in the UK Biobank cohort are presented in Table S9 in the online supplement; this analysis revealed 12 independent GWAS signals located in eight loci. The UK Biobank GWASs of AUDIT-C and AUDIT-P score subsets are summarized in Tables S10 and S11 and Figure S4 in the online supplement. Seven of these 12 independent GWAS signals were also significantly associated with AUDIT-C score; the same index variants were identified in the two analyses. An additional GWAS signal was also identified close to FNBP4. For AUDIT-P score, five independent GWAS signals were significantly associated, and these loci were also associated with the total AUDIT and AUDIT-C scores. The rs1229984 SNP in ADH1B was not available for meta-analysis in the 23andMe sample and was not in Hardy-Weinberg equilibrium in the UK Biobank sample used in the present study (p=3.2×10−16); however, in the total UK Biobank white British sample, there was no significant deviation from Hardy-Weinberg equilibrium (p=0.13). The associations between rs1229984 and AUDIT scores are presented in Tables S9, S10, and S11 in the online supplement. The rs1229984 SNP was strongly associated with all AUDIT scores in the UK Biobank sample (β=0.04–0.06, p≤1.0×10−45), but this SNP was not included for clump-based pruning and downstream analyses. We therefore performed a conditional analysis of the SNPs on 4q23 and 4q24 in the UK Biobank sample to determine whether any of these associations were significant after controlling for rs1229984 genotype. While rs13107325 on 4q24 remained significantly associated with AUDIT total score after controlling for rs1229984 genotype, the association between rs146788033, rs11733695, and rs3114045 and AUDIT total score became nonsignificant, suggesting that these loci are tagging the strong rs1229984 signal in this region.
GWAS Meta-Analysis of AUDIT Total Score
The GWAS meta-analysis of the UK Biobank and 23andMe samples found 15 independent GWAS signals (see Table S12 in the online supplement) associated with AUDIT total score spanning 10 genomic loci (Table 1). Figure 1 presents the Manhattan and quantile-quantile plots of the GWAS meta-analysis of AUDIT total score, and Figures S5–S14 in the online supplement present the regional association plots for the independent signals. The inflation factor of the meta-analysis GWAS was λGC=1.22 with an LDSC intercept of 1.008 (SE=0.007), suggesting that the majority of the inflation is due to polygenicity. The 15 independent SNPs show 100% sign concordance for association with AUDIT total score across the UK Biobank and 23andMe samples (Table 1); 11 of these SNPs were nominally associated with AUDIT total score in the 23andMe sample (p≤0.05), and all index SNPs were associated with AUDIT total score in the UK Biobank sample (p<1.8×10−6).
| UK Biobank (N=121,604) | 23andMe (N=20,328) | Meta-Analysis (N=141,932) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chromosome | Index Variant | Genes (Number of Credible SNPs) | A1 | Beta | SE | p | A1 | Beta | SE | p | A1 | Z Score | p | Direction |
| 2p21 | rs4953148 | LINC01833 (17) | T | 0.007 | 0.001 | 2.8×10–8 | T | 0.03 | 0.036 | 3.5×10–1 | A | –5.59 | 2.3×10–8 | −/− |
| 2p23.3 | rs1260326 | SNX17 (1), GCKR (3) | C | 0.008 | 0.001 | 2.1×10–10 | T | –0.10 | 0.034 | 2.7×10–3 | T | –7.15 | 8.7×10–13 | −/− |
| 3q25.33 | rs1920650 | SMC4, TRIM59, B3GAT3P1, KPNA4, SCARNA7, KRT8P12, RPL6P8, ARL14 (132) | C | 0.006 | 0.001 | 1.0×10–6 | T | –0.07 | 0.037 | 4.2×10–2 | T | –5.63 | 1.7×10–8 | −/− |
| 4p14 | rs11940694 | KLB (7) | G | 0.01 | 0.001 | 9.8×10–20 | G | 0.08 | 0.034 | 2.5×10–2 | A | –9.38 | 6.8×10–21 | −/− |
| 4p14 | rs4975012 | KLB (6) | A | 0.008 | 0.001 | 1.9×10–8 | G | –0.006 | 0.003 | 4.9×10–2 | A | 5.94 | 2.8×10–9 | +/+ |
| 4q23 | rs146788033 | METAP1 (1) | G | –0.04 | 0.005 | 1.5×10–22 | G | –0.03 | 0.008 | 1.4×10–4 | A | 10.48 | 1.0×10–25 | +/+ |
| 4q23 | rs11733695 | RP11–696N14.1 (1) | A | –0.07 | 0.006 | 9.1×10–26 | G | 0.05 | 0.012 | 6.7×10–6 | A | –11.42 | 3.4×10–30 | −/− |
| 4q23 | rs3114045 | ADH1C (12) | C | 0.01 | 0.002 | 3.4×10–10 | T | –0.15 | 0.048 | 1.6×10–3 | T | –7.18 | 7.2×10–13 | −/− |
| 4q24 | rs188514326 | RP11–588P8.1 (1) | C | –0.04 | 0.009 | 3.0×10–7 | G | 0.03 | 0.013 | 4.9×10–2 | C | –5.49 | 4.1×10–8 | −/− |
| 4q24 | rs13135092 | RN7SL728P (1), SLC39A8 (5) | G | –0.02 | 0.002 | 4.1×10–13 | G | –0.09 | 0.060 | 1.2×10–1 | A | 7.59 | 3.2×10–14 | +/+ |
| 8q22.1 | rs35040843 | RP11–700E23.3 (30) | T | 0.007 | 0.001 | 1.7×10–7 | T | 0.09 | 0.038 | 8.9×10–3 | T | 5.55 | 2.9×10–8 | +/+ |
| 10p11.23 | rs7078436 | JCAD (9) | G | –0.006 | 0.001 | 1.8×10–6 | G | –0.10 | 0.035 | 3.1×10–3 | A | 5.74 | 9.6×10–9 | +/+ |
| 11p11.2 | rs2293576 | NDUFS3 (3), SPI1 (6), SLC39A13 (3), PSMC3 (3), RAPSN (6), CELF1 (16), RP11–750H9.5 (15), Y_RNA (1) | A | 0.007 | 0.001 | 5.0×10–9 | G | –0.06 | 0.035 | 1.2×10–1 | A | 5.83 | 5.5×10–9 | +/+ |
| 17q21.31 | rs62062288 | CRHR1 (142), DND1P1 (1), ENSG00000262372 (1), ENSG00000262500 (2), ENSG00000262881 (12), KANSL1 (376), MAPT (755), MAPT-AS1 (255), NSF (15), PLEKHM1 (1), SPPL2C (11), STH (1), RP11–105N13.4 (541), WNT3 (16) | A | –0.009 | 0.001 | 1.6×10–9 | G | 0.09 | 0.042 | 2.1×10–2 | A | –6.21 | 5.4×10–10 | −/− |
| 19q13.33 | rs492602 | FUT2 (29), MAMSTR (8), RASIP1 (23), IZUMO1 (6) | G | 0.006 | 0.001 | 7.0×10–8 | G | 0.05 | 0.033 | 1.7×10–1 | A | –5.57 | 2.5×10–8 | −/− |
TABLE 1. Index SNPs Associated With AUDIT Scores in a GWAS Meta-Analysis of AUDIT Scores and Corresponding p Values in the UK Biobank and 23andMe Cohortsa
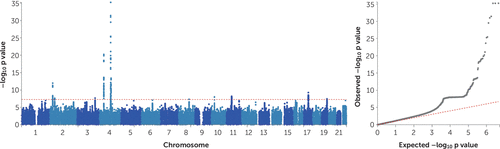
FIGURE 1. Manhattan and QQ Plots for the SNP-Based GWAS Meta-Analysis of AUDIT Total Score (N=141,932)a
a AUDIT=Alcohol Use Disorders Identification Test; GWAS=genome-wide association study; QQ=quantile-quantile; SNP=single-nucleotide polymorphism.
The top hit for the GWAS meta-analysis of AUDIT total score was a variant (rs11733695) located downstream (879 base pairs) from ADH6 (p=3.4×10−30), which is a member of the alcohol dehydrogenase gene family. The rs11733695 SNP is in low LD (r2=0.17) with the functional SNP in ADH1B, rs1229984, which is known to alter alcohol metabolism (26). In addition, two other regions in 4q23 were associated with AUDIT total score in the meta-analysis: one of the index SNPs was located in the ADH1C gene; however, conditional analysis of this region in the UK Biobank sample alone suggests that these multiple hits may in fact be tagging the rs1229984 signal. This region has been previously associated with alcohol consumption, alcohol use disorders, and AUDIT scores (6, 7, 9, 10).
We also replicated the association between KLB (see Table S12 in the online supplement), on chromosome 4q14, and alcohol consumption (9, 11, 12); the index SNP rs11940694, which is located in the intron of KLB, was associated with AUDIT total score in the present study. Clump-based pruning identified rs11940694 and rs4975012 as independent hits in the KLB region. Credible set analysis suggests that rs2046330 is the index SNP in the region represented by rs4975012 (see Table S13 in the online supplement). AUDIT total score was also associated with SNPs that localized to GCKR on chromosome 2p23.3, which has been previously associated with alcohol consumption (9, 11). Five SNPs comprised the credible set at the GCKR locus, which also spans the SNX17 gene, including the missense variant (rs1260326) in GCKR that was identified as the index SNP.
We identified GWAS signals in several regions that have not been previously implicated in the genetics of alcohol use disorders, including 2p21, 17q21, 3q25, 8q22, 10p11, 11p11, and 19q13. The index SNP rs13135092 in the 4q24 region is located in an intron of SLC39A8; the remainder of the credible set for this locus is located in a noncoding pseudogene, RN7SL728P. SLC39A8 is highly pleiotropic (27), but it is a novel association in relation to alcohol. A region of association on 2p21 contains 17 SNPs that are localized to the noncoding RNA LINC01833. A novel region of association was also detected on chromosome 10p11.23; this region contains nine credible SNPs that localize to the JCAD (junctional cadherin 5 associated) gene. JCAD encodes an endothelial cell junction protein and has previously been associated with coronary heart disease (28).
The remaining novel associations on 3q25, 8q, 11p11, 17q21, and 19q13.3 were more complex. The index variants on chromosome 8q22.1 were not localized to any genes, and it is unclear from the credible set analyses what the causal variants may be at these loci. The credible SNP sets for the 3q25.33, 11p11.2, 17q21.31, and 19q13.3 regions contained more than 50 SNPs each, which spanned several genes. For example, the index SNP on chromosome 17q21.31 was an intronic SNP in MAPT, which encodes the tau protein and has been robustly associated with Parkinson’s disease (30, 31) (see Table S14 in the online supplement) and other neurodegenerative tauopathies (32) and, more recently, with neuroticism (33). However, we note that the region of association on chromosome 17q21.31 spans the corticotropin receptor gene (CRHR1), which has been associated with alcohol use in animals and humans (34). Thus, because of the extended complex LD in this region, we are unable to determine the likely causal variant. Similarly, the index SNP (rs2293576) at chromosome 11p11.2 is a synonymous SNP of the zinc transporter gene SLC39A13; however, this region includes 54 associated SNPs, which map to five additional genes. Lastly, the index SNP on 19q13.3 is a synonymous variant in FUT2. FUT2 encodes galactoside 2-alpha-l-fucosyltransferase 2, which controls the expression of ABO blood group antigens. However, 70 SNPs form this credible set, and they span four genes in total.
We used FUMA to functionally annotate all 1,290 SNPs in the credible sets (see Table S13 in the online supplement). The majority of the SNPs were intronic (76.9%; N=993) or intergenic (11.6%; N=149), and only 26 SNPs (2.0%) were exonic. Furthermore, 38 SNPs showed CADD scores >12.37, which is the suggested threshold to be considered deleterious (35). The exonic SNPs (rs601338, rs17651549, rs13107325) of FUT2, MAPT, and SLC39A8, respectively, had the highest CADD scores (>34), suggesting potential deleterious protein effects. Overall, 164 SNPs had RegulomeDB scores of 1a−1f, showing evidence of potential regulatory effects; 90.1% of the SNPs were in open chromatin regions (minimum chromatin state, 1–7).
Gene-Based and Pathway Analyses
We used MAGMA (22) to perform a gene-based association analysis, which identified 40 genes that were significantly associated with AUDIT total score (p<2.70×10−6) (see Table S15 and Figure S15 in the online supplement). As expected, the majority of these genes were in the 10 GWAS loci (i.e., KLB, GCKR); DRD2 and CRHR1 were also among the top hits. In addition, the analysis revealed a strong burden signal in CADM2 (p=1.64×10−9), where the index variant in GWAS meta-analysis did not reach genome-wide significance. We did not identify any canonical pathways that were significantly associated with AUDIT score (see Table S16 in the online supplement).
Gene-based (MAGMA) analyses for the AUDIT-C and AUDIT-P subsets (see Figures S16 and S17 in the online supplement) revealed evidence of overlap (see Figure S18 and Table S17 in the online supplement). Two genes (KLB, CADM2) were associated with all three AUDIT traits (AUDIT total score, AUDIT-C score, and AUDIT-P score). There was considerable overlap between AUDIT total score and AUDIT-C score, with 21 overlapping genes associated at the gene-based level. Only one gene, DRD2, was associated with both AUDIT total score and AUDIT-P score.
S-PrediXcan
S-PrediXcan identified a positive correlation (p<1.07×10−6) between AUDIT total score and the predicted expression of 26 genes across multiple brain tissues (full results are presented in Table S18 in the online supplement), including MAPT (cerebellum), FUT2 (caudate and nucleus accumbens), and CRHR1-IT1 (putamen, cerebellum, hippocampus, anterior cingulate cortex). SNPs in the region of MAPT and FUT2 were associated with AUDIT total score in the GWAS. MAPT (cerebellum), FUT2 (nucleus accumbens), and CRHR1-IT1 (caudate, cortex, nucleus accumbens, hypothalamus, hippocampus) were also associated with AUDIT-C score (see Table S19 in the online supplement). S-PrediXcan for AUDIT-C and AUDIT-P scores (see Table S20 in the online supplement) revealed that lower predicted RFC1 expression in the cerebellum and hemisphere, respectively, was associated with higher scores on both the AUDIT-C (p=7.84×10−7) and the AUDIT-P (p=1.54×10−6).
Genetic Correlations
We used LDSC to evaluate evidence for genetic correlations between our three primary traits (AUDIT total score, AUDIT-C score, and AUDIT-P score) and numerous other traits for which GWAS summary statistics were available; these included alcohol and substance use traits, personality and behavioral traits, psychiatric disorders, blood lipid levels, and brain structure volumes (see Figure 2; see also Tables S3–S5 in the online supplement).
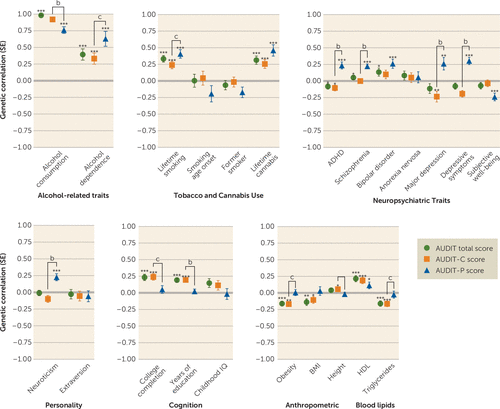
FIGURE 2. Genetic Correlations Between the Three AUDIT Phenotypes (Total Score, AUDIT-C Score, AUDIT-P Score) and Several Traits Measured in Independent Cohortsa
a More information on the various traits is provided in Tables S3–S5 in the online supplement. ADHD=attention deficit hyperactivity disorder; AUDIT=Alcohol Use Disorders Identification Test; AUDIT-C=AUDIT items 1–3 (consumption); AUDIT-P=AUDIT items 4–10 (problematic consequences of drinking); BMI=body mass index.
b AUDIT-P compared with AUDIT-C, p<0.01, false discovery rate 5%.
c AUDIT-P compared with AUDIT-C, p<0.05.
*p<0.05. **p<0.01. ***p<0.0001.
As expected, AUDIT-C and AUDIT-P scores were highly genetically correlated (rg=0.70, p=4.1×10−70). AUDIT scores (AUDIT total score, AUDIT-C score, and AUDIT-P score) showed strong genetic correlations with alcohol consumption from two other studies (rg=0.76–0.96, p<2.3×10−9). Many of the genetic correlations with AUDIT-P score were significantly different from the correlations with AUDIT-C score (see Table S6 in the online supplement). AUDIT-C score had a significantly stronger (p=8.02×10−3) correlation with alcohol consumption (rg=0.92, p=7.0×10−164) than did AUDIT-P score (rg=0.76, p=2.7×10−52). In contrast, AUDIT total score and AUDIT-C score were only modestly correlated with alcohol dependence (rg=0.39 and 0.33, respectively, p<8.2×10−5), whereas AUDIT-P score showed a nominally stronger genetic correlation with alcohol dependence (rg=0.63, p=1.8×10−8; AUDIT-P compared with AUDIT-C, p=0.033) (see Table S6).
We detected positive genetic correlations between AUDIT scores (AUDIT total, AUDIT-C score, AUDIT-P score) and other substance use phenotypes, including lifetime smoking (rg=0.24–0.41, p<1.6×10−5) and cannabis use (rg=0.26–0.46, p<1.1×10−4). We also observed a positive genetic correlation between AUDIT-P score and cigarettes per day (rg=0.28, p=4.0×10−3).
Several psychiatric disorders and related traits were positively genetically correlated with AUDIT-P score, including schizophrenia (rg=0.22, p=3.0×10−10), bipolar disorder (rg=0.26, p=1.5×10−4), ADHD (rg=0.23, p=1.1×10−5), and major depressive disorder (rg=0.26, p=50.6×10−3). Intriguingly, AUDIT-C score was negatively correlated with major depressive disorder (rg=−0.23, p=3.7×10−3) and ADHD (rg=−0.10, p=1.8×10−2), whereas AUDIT-P score showed positive genetic correlations with these same disease traits (rg=0.26, p=5.6×10−3; rg=0.24, p=1.1×10−5). We observed a positive genetic correlation between AUDIT-P score and depressive symptoms (rg=0.30, p=3.0×10−8) and neuroticism (rg=0.18, p=2.6×10−4) and a negative genetic correlation with subjective well-being (rg=−0.24, p=4.0×10−5).
We observed positive genetic correlations between AUDIT total score and AUDIT-C score and education, college completion, and cognitive ability (rg=0.19–0.24, p<1.5×10−5). The genetic correlations between AUDIT-P and education and college completion were near zero and were significantly lower than AUDIT-C score and AUDIT total score or education traits (see Table S6 in the online supplement).
There were negative genetic correlations between AUDIT total score and AUDIT-C score and obesity (rg=−0.16–0.17, p<1.1×10−5), similar to previous reports regarding AUDIT total score (10) and alcohol consumption (9). In contrast, obesity was not significantly genetically correlated with AUDIT-P score (rg=0.01, p=0.90). Similarly, HDL cholesterol and triglyceride levels were genetically correlated with AUDIT total score and AUDIT-C score (rg=0.19–22, p<9.3×10−5, rg=−0.16, p<1.0×10−4, respectively), but this association was not found for AUDIT-P score (rg=0.11, p=2.2×10−2, rg=−0.03, p=6.4×10−1). Obesity showed significantly different correlations with both AUDIT-P and AUDIT-C scores (see Table S6).
Dichotomizing AUDIT Total Score to More Closely Approximate DSM-IV Alcohol Dependence
As AUDIT can be rapidly ascertained in large populations, we explored methods for dichotomizing AUDIT total score to optimize the genetic correlation with DSM-IV alcohol dependence (13). Higher genetic correlations with alcohol dependence were observed as the control threshold was increased from 2 to 4, and with increasingly stringent case cutoffs (Figure 3; see also Table S2 in the online supplement). The highest genetic correlation was observed for cases with AUDIT total score ≥20 and controls ≤4 (rg=0.90, SE=0.25, p=3.0×10−4); however, this highly stringent threshold produced very few cases (N=1,290). The standard error of the estimate is much larger at more stringent case thresholds, and therefore these estimates should be interpreted with caution. Defining cases as ≥12 yielded an rg of 0.82 (SE=0.18, p=3.2×10−6) while retaining more than seven times as many cases (N=9,130); these genetic correlations were not significantly different from those obtained using cases ≥20 and controls ≤4 (p=0.80).
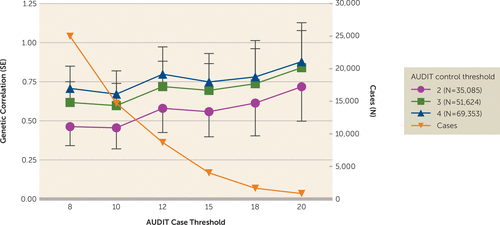
FIGURE 3. Dichotomizing AUDIT Total Score to Approximate DSM-IV Alcohol Dependencea
a Continuous AUDIT total score was used to categorize participants as case and control subjects at different thresholds; GWAS was performed on each, and the genetic correlation with DSM-IV alcohol dependence was calculated. Thresholds for AUDIT-determined cases were 8 (N=25,423), 10 (N=15,151), 12 (N=9,130), 15 (N=4,471), 18 (N=2,099), and 20 (N=1,290). The orange line is a visualization of the number of cases used at each threshold, corresponding to the N on the right-hand y-axis. AUDIT=Alcohol Use Disorders Identification Test; GWAS=genome-wide association study.
Discussion
We have presented the largest GWAS meta-analysis of AUDIT total score to date, using large population-based cohorts from the UK Biobank and 23andMe. We identified novel associations with AUDIT total score; the genes located in these regions include JCAD and SLC39A8. We found evidence for association in several loci previously associated with alcohol use via single-variant and gene-based analyses (i.e., KLB, GCKR, CADM2). The SNP heritability of all AUDIT phenotypes ranged from 9% to 12%, demonstrating that common genetic factors account for a prominent proportion of the variation in alcohol use phenotypes. Furthermore, we showed that there is shared genetic architecture between AUDIT scores and other alcohol and substance use phenotypes. AUDIT-P score showed a positive genetic correlation with several psychiatric diseases, distinguishing AUDIT-P score from AUDIT-C score. Finally, using LDSC, we identified thresholds for dichotomizing AUDIT total score (score ≥12 to define cases and ≤4 to define controls) that maximize the genetic correlation with alcohol dependence while retaining a large number of participants.
Our top GWAS hits replicated previous association signals for alcohol use traits. The strongest associations with AUDIT score in this study spanned the alcohol metabolism genes on chromosome 4q23 (10). Variants in this region were associated with AUDIT total score, AUDIT-C score, and AUDIT-P score, demonstrating that alcohol metabolism is a risk factor for both alcohol consumption and problematic use. The second strongest signal, also associated with the three AUDIT phenotypes, is located in KLB, confirming the robust association of this gene with alcohol consumption in both humans (9, 11, 12) and mice (12). However, the biology of this locus could be more complex than previously described. Although the credible set analysis suggested that the more probable causal variants are all located on the first intron of KLB, one of these variants, rs11940694, is an expression quantitative trait locus for RFC1 expression in the brain, and S-PrediXcan analysis predicted that lower expression of RFC1 in the brain is associated with higher predicted AUDIT (AUDIT-C and AUDIT-P) scores. Interestingly, a gene in the complex GWAS signal on chromosome 19, Fibroblast growth factor 21 (FGF21), was associated with AUDIT scores (AUDIT total score, AUDIT-C score, AUDIT-P score) at the gene-based level (see Table S17 in the online supplement). FGF21 regulates sweet and alcohol preference in mice as part of a receptor complex with β-Klotho (KLB) in the central nervous system (33). Additionally, we replicated the association between rs1260326 in the gene GCKR and alcohol consumption (9, 11), here associated with AUDIT total score and AUDIT-C score. Other loci previously associated with alcohol consumption include CADM2 (9), which was associated at the gene-based level for all three AUDIT traits. Here, the burden analysis suggests that multiple (rare and common) variants are necessary to explain the association signal. Intriguingly, several of the novel associations with AUDIT scores were mapped to highly pleiotropic genes (MAPT, FUT2, SLC39A8) (27).
Genetic analysis of the AUDIT subsets revealed evidence of distinct genetic architecture between AUDIT-C and AUDIT-P (alcohol consumption versus problem use), with support from the gene-based (see Figures S16 and S17 in the online supplement), S-PrediXcan (see Tables S19 and S20 in the online supplement), and genetic correlation analyses (see Figure 2). Furthermore, AUDIT-P score showed a strong genetic correlation with alcohol dependence (13). In contrast, AUDIT-C score had a stronger genetic correlation with alcohol consumption. Thus, partitioning AUDIT scores into different subsets (alcohol consumption versus problem use) may disentangle genetic factors that contribute to different aspects of vulnerability to alcohol use disorders.
Genetic overlap was observed for all measures of AUDIT and other substance use traits, including lifetime tobacco and cannabis use, as we previously reported (10, 36, 37), demonstrating that genetic risk factors for high AUDIT scores overlap with increased consumption of multiple drug types.
We found several significant differences between the genetic correlations with AUDIT-P and AUDIT-C scores. These differences were particularly pronounced for psychiatric and behavioral traits. AUDIT-P score was positively genetically correlated with psychopathology (schizophrenia, bipolar disorder, major depressive disorder, ADHD), personality traits (including neuroticism), and regional brain volumes. These associations have previously been observed at the phenotypic level; alcohol use disorders commonly co-occur in individuals with schizophrenia (38), bipolar disorder (39), major depressive disorder (40), and adult ADHD (41). Intriguingly, genetic risk for high AUDIT-C score was negatively correlated with major depressive disorder and ADHD, demonstrating that a distinct genetic component of AUDIT-P is shared with genetic risk for psychiatric disease. Regional volume abnormalities in subcortical brain regions of individuals with alcohol use disorders have been reported (42–44), although it is unclear whether these alterations are a result of high alcohol consumption or a preexisting susceptibility. We identified a positive genetic correlation between AUDIT-P score and increased caudate volume; however, the majority of studies report reductions in regional brain volumes associated with alcohol use disorders.
For AUDIT total score and AUDIT-C score, we showed positive genetic correlations with educational attainment and cognitive ability and negative genetic correlations with obesity, consistent with earlier reports (9, 10). These associations were not observed for AUDIT-P score. Similarly, HDL cholesterol levels showed a significant positive correlation, and triglyceride levels a negative correlation, with AUDIT total score and AUDIT-C score, but not AUDIT-P score. These patterns were previously observed for alcohol consumption (9). We could speculate that these differences may be linked to socioeconomic status. Alcohol consumption is often higher in individuals with higher socioeconomic status (45), whereas alcohol-related problems, such as binge drinking (46) and alcohol-related mortality (47), are more prevalent in individuals with lower socioeconomic status. Furthermore, individuals with low socioeconomic status are more likely to have alcohol use disorders with psychiatric comorbidities (48). Consistent with this idea, we find positive genetic correlations between AUDIT-C score and education, a trait correlated with socioeconomic status (49), and positive genetic correlations between AUDIT-P score and psychopathology. Our findings provide further evidence that different dimensions of alcohol use associate differently with behavior and that these differences may have a biological underpinning.
A clinical diagnosis of an alcohol use disorder is often required to define cases for genetic studies. An alternative strategy would be to use AUDIT to infer alcohol use disorder case status; however, it has not been clear whether and how to perform meta-analyses between AUDIT scores and alcohol dependence. A GWAS meta-analysis for AUDIT score and alcohol dependence could be simplified if a threshold could be used to define cases and controls based on AUDIT score, an approach that was used by Mbarek et al. (50). We have provided empirical evidence about genetic correlations between AUDIT score and alcohol dependence using dichotomized AUDIT scores and found thresholds for AUDIT score that produced high genetic correlations with alcohol use disorders (see Figure 3). Genetic correlations increased as the upper threshold for cases was made more stringent, although the standard errors for all of these estimates were overlapping. The genetic correlation with alcohol dependence appeared to become asymptotic when case status was defined as ≥12; therefore, this threshold could be used to define case status. We also considered various thresholds for defining controls and found that a threshold of ≤4 produced a high genetic correlation with alcohol dependence while also retaining the largest number of subjects.
Our study is not without limitations. The AUDIT specifically asks about the past year, and thus it may not capture information on lifetime alcohol use and misuse. This is suboptimal for genetic studies, because it effectively measures a recent state rather than a stable trait. Measures capturing drinking and alcohol use disorders across the lifespan may be preferable. Also, although the mean AUDIT-C score was 4.24, the mean AUDIT-P score was considerably lower (0.75). Thus, we were not able to perform a more refined categorization (e.g., three subsets: consumption [items 1–3], dependence [items 4–6], hazardous use [items 7–10]), as fewer individuals endorsed the items comprising the AUDIT-P (see Table S7 in the online supplement). Furthermore, our study uses data from the UK Biobank and 23andMe research participants, who were volunteers not ascertained for alcohol use disorders, and hence our findings may not generalize to populations that show higher rates of alcohol use and dependence. Additional alcohol-related phenotypes (e.g., age at first use and patterns of alcohol drinking, including binge drinking) could be used in subsequent genetic studies to identify additional sources of genetic vulnerability for alcohol use disorders. Lastly, we offered guidelines to identify cases to use in genetic studies of alcohol use disorders (i.e., an AUDIT score ≥12) based on genetic correlations; however, these recommendations are not intended to determine thresholds for diagnosing dependence in a clinical setting. Future studies will be able to test whether using the AUDIT as a surrogate for alcohol use disorders will be beneficial for gene discovery. In addition, several studies have argued that lower thresholds should be used for females, which has not been addressed in the present study.
Conclusions
We have reported the largest GWAS of AUDIT ever undertaken. We replicated previously identified signals (i.e., ADH1B, ADH1C; KLB; GCKR) and identified novel GWAS signals (i.e., JACD, SLC39A8) associated with AUDIT scores. We showed that different portions of the AUDIT (AUDIT-C, AUDIT-P) correlate with distinct traits, which will aid in dissecting genetic vulnerability to alcohol use and dependence. The genetic factors that predispose to high alcohol consumption inevitably overlap with those for problem drinking, as heavy drinking is generally a prerequisite for the development of hazardous use and dependence. However, not everyone who consumes alcohol experiences the same level of harmful consequences. By studying the different subsets of AUDIT, we identified genetic factors that may be specific to problem drinking. Larger studies of cohorts with a wider range of AUDIT-P scores are required to both replicate and expand these findings. Finally, we described an alternative strategy to rigorous ascertainment for genetic studies of alcohol use disorders, namely, an AUDIT score ≥12 to define cases and a score ≤4 to define controls, which could be used to achieve large sample sizes in a cost-efficient manner.
1 : Genetic differences in alcohol sensitivity and the inheritance of alcoholism risk. Psychol Med 1999; 29:1069–1081Crossref, Medline, Google Scholar
2 : The heritability of alcohol use disorders: a meta-analysis of twin and adoption studies. Psychol Med 2015; 45:1061–1072Crossref, Medline, Google Scholar
3 : Genome-wide significant association between alcohol dependence and a variant in the ADH gene cluster. Addict Biol 2012; 17:171–180Crossref, Medline, Google Scholar
4 : Alcohol consumption mediates the relationship between ADH1B and DSM-IV alcohol use disorder and criteria. J Stud Alcohol Drugs 2014; 75:635–642Crossref, Medline, Google Scholar
5 : ALDH2 is associated to alcohol dependence and is the major genetic determinant of “daily maximum drinks” in a GWAS study of an isolated rural Chinese sample. Am J Med Genet B Neuropsychiatr Genet 2014; 165B:103–110Crossref, Medline, Google Scholar
6 : Genome-wide association study of alcohol dependence: significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry 2014; 19:41–49Crossref, Medline, Google Scholar
7 : ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol Psychiatry 2012; 17:445–450Crossref, Medline, Google Scholar
8 : Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption, and dependence: an integrated analysis. Hum Mol Genet 2009; 18:580–593Crossref, Medline, Google Scholar
9 : Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112 117). Mol Psychiatry 2017; 22:1376–1384Crossref, Medline, Google Scholar
10 : Genome-wide association study of Alcohol Use Disorder Identification Test (AUDIT) scores in 20 328 research participants of European ancestry. Addict Biol (Epub ahead of print, Oct 23, 2017)Google Scholar
11 : Genetic contributors to variation in alcohol consumption vary by race/ethnicity in a large multi-ethnic genome-wide association study. Mol Psychiatry 2017; 22:1359–1367Crossref, Medline, Google Scholar
12 : KLB is associated with alcohol drinking, and its gene product β-Klotho is necessary for FGF21 regulation of alcohol preference. Proc Natl Acad Sci USA 2016; 113:14372–14377Crossref, Medline, Google Scholar
13 : Trans-ancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. bioRxiv 257311; March 10, 2018 (doi:
14 : AUDIT questionnaire. Addiction 1993; 88:1–2Google Scholar
15 : The Alcohol Use Disorders Identification Test: Guidelines for Use in Primary Care, 2nd ed. Geneva, World Health Organization, 2001Google Scholar
16 : LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015; 47:291–295Crossref, Medline, Google Scholar
17 : UK Biobank: current status and what it means for epidemiology. Health Policy Technol 2012; 1:123–126Crossref, Google Scholar
18 : Genome-wide genetic data on ∼500,000 UK Biobank participants. bioRxiv 166298; July 20, 2017 (https://www.biorxiv.org/content/early/2017/07/20/166298)Google Scholar
19 : GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011; 88:76–82Crossref, Medline, Google Scholar
20 : METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26:2190–2191Crossref, Medline, Google Scholar
21 : Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet 2012; 44:1294–1301Crossref, Medline, Google Scholar
22 : Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017; 8:1826Crossref, Medline, Google Scholar
23 : Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. bioRxiv 045260; March 23, 2016 (https://www.biorxiv.org/content/early/2017/10/03/045260)Google Scholar
24 : An atlas of genetic correlations across human diseases and traits. Nat Genet 2015; 47:1236–1241Crossref, Medline, Google Scholar
25 : Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995; 57:289–300Google Scholar
26 : Genes encoding enzymes involved in ethanol metabolism. Alcohol Res 2012; 34:339–344Medline, Google Scholar
27 : Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet 2016; 48:709–717Crossref, Medline, Google Scholar
28 : Genome-wide association study identifies a new locus for coronary artery disease on chromosome 10p11.23. Eur Heart J 2011; 32:158–168Crossref, Medline, Google Scholar
29 : A genome-wide association meta-analysis of diarrhoeal disease in young children identifies FUT2 locus and provides plausible biological pathways. Hum Mol Genet 2016; 25:4127–4142Crossref, Medline, Google Scholar
30 : Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet 2009; 41:1303–1307Crossref, Medline, Google Scholar
31 : Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet 2009; 41:1308–1312Crossref, Medline, Google Scholar
32 : Untangling the tau gene association with neurodegenerative disorders. Hum Mol Genet 2006; 15:R188–R195Crossref, Medline, Google Scholar
33 : FGF21 regulates sweet and alcohol preference. Cell Metab 2016; 23:344–349Crossref, Medline, Google Scholar
34 : Corticotropin releasing factor: a key role in the neurobiology of addiction. Front Neuroendocrinol 2014; 35:234–244Crossref, Medline, Google Scholar
35 : A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014; 46:310–315Crossref, Medline, Google Scholar
36 : Connecting the dots: genome-wide association studies in substance use. Mol Psychiatry 2016; 21:733–735Crossref, Medline, Google Scholar
37 : Polygenic risk scores for smoking: predictors for alcohol and cannabis use? Addiction 2014; 109:1141–1151Crossref, Medline, Google Scholar
38 : Co-occurring alcohol use disorder and schizophrenia. Alcohol Res Health 2002; 26:99–102Google Scholar
39 : Substance abuse and bipolar comorbidity. Psychiatr Clin North Am 1999; 22:609–627, ixCrossref, Medline, Google Scholar
40 : Comorbidity of alcoholism and psychiatric disorders: an overview. Alcohol Res Health 2002; 26:81–89Google Scholar
41 : Comorbidity of alcohol and substance dependence with attention-deficit/hyperactivity disorder (ADHD). Alcohol Alcohol 2008; 43:300–304Crossref, Medline, Google Scholar
42 : Longitudinal changes in magnetic resonance imaging brain volumes in abstinent and relapsed alcoholics. Alcohol Clin Exp Res 1995; 19:1177–1191Crossref, Medline, Google Scholar
43 : Brain gray and white matter volume loss accelerates with aging in chronic alcoholics: a quantitative MRI study. Alcohol Clin Exp Res 1992; 16:1078–1089Crossref, Medline, Google Scholar
44 : Chronic active heavy drinking and family history of problem drinking modulate regional brain tissue volumes. Psychiatry Res 2005; 138:115–130Crossref, Medline, Google Scholar
45 : Associations between socioeconomic factors and alcohol outcomes. Alcohol Res 2016; 38:83–94Medline, Google Scholar
46 : Socioeconomic patterning of excess alcohol consumption and binge drinking: a cross-sectional study of multilevel associations with neighbourhood deprivation. BMJ Open 2013; 3:3Crossref, Google Scholar
47 : Socioeconomic differences in alcohol-attributable mortality compared with all-cause mortality: a systematic review and meta-analysis. Int J Epidemiol 2014; 43:1314–1327Crossref, Medline, Google Scholar
48 : Functional limitations, socioeconomic status, and all-cause mortality in moderate alcohol drinkers. J Am Geriatr Soc 2009; 57:955–962Crossref, Medline, Google Scholar
49 : Molecular genetic contributions to socioeconomic status and intelligence. Intelligence 2014; 44:26–32Crossref, Medline, Google Scholar
50 : The genetics of alcohol dependence: twin and SNP-based heritability, and genome-wide association study based on AUDIT scores. Am J Med Genet B Neuropsychiatr Genet 2015; 168:739–748Crossref, Medline, Google Scholar



