Polygenic Risk Score for Alzheimer’s Disease: Implications for Memory Performance and Hippocampal Volumes in Early Life
Abstract
Objective:
Alzheimer’s disease is a heritable neurodegenerative disorder in which early-life precursors may manifest in cognition and brain structure. The authors evaluate this possibility by examining, in youths, associations among polygenic risk score for Alzheimer’s disease, cognitive abilities, and hippocampal volume.
Method:
Participants were children 6–14 years of age in two Brazilian cities, constituting the discovery (N=364) and replication samples (N=352). As an additional replication, data from a Canadian sample (N=1,029), with distinct tasks, MRI protocol, and genetic risk, were included. Cognitive tests quantified memory and executive function. Reading and writing abilities were assessed by standardized tests. Hippocampal volumes were derived from the Multiple Automatically Generated Templates (MAGeT) multi-atlas segmentation brain algorithm. Genetic risk for Alzheimer’s disease was quantified using summary statistics from the International Genomics of Alzheimer’s Project.
Results:
Analyses showed that for the Brazilian discovery sample, each one-unit increase in z-score for Alzheimer’s polygenic risk score significantly predicted a 0.185 decrement in z-score for immediate recall and a 0.282 decrement for delayed recall. Findings were similar for the Brazilian replication sample (immediate and delayed recall, β=−0.259 and β=−0.232, both significant). Quantile regressions showed lower hippocampal volumes bilaterally for individuals with high polygenic risk scores. Associations fell short of significance for the Canadian sample.
Conclusions:
Genetic risk for Alzheimer’s disease may affect early-life cognition and hippocampal volumes, as shown in two independent samples. These data support previous evidence that some forms of late-life dementia may represent developmental conditions with roots in childhood. This result may vary depending on a sample’s genetic risk and may be specific to some types of memory tasks.
Alzheimer’s disease is a late-life neurodegenerative disorder affecting 1 in 9 Americans over age 65 (1). No effective treatments exist, perhaps because Alzheimer’s disease is diagnosed relatively late in a slowly evolving disease process (2, 3). In fact, evidence suggests that this process begins in early life (4), much like other chronic disorders of adults, including obesity and cardiovascular disease. The present study evaluated early-life signs of developmental risk for Alzheimer’s disease by examining associations among genetic risk, cognition, and brain structure in children and adolescents.
Heritability in Alzheimer’s disease has been estimated to be in the range of 48%–79% (5, 6). Studies of specific genetic risk factors have identified the apolipoprotein E (APOE) ε4 allele (7) and multiple other risk factors of smaller effect size detected in genome-wide association studies (GWAS) (8–13). Even though these variants have a low impact on Alzheimer’s disease risk individually, the additive effect of several loci may have significant predictive utility (14). The sum of the alleles associated with a certain trait for a given p value threshold, weighted by their effect sizes estimated from GWAS, is called polygenic risk score.
Evidence of a slowly evolving course in Alzheimer’s disease arises from various sources. Neuropathological features can appear decades before disease onset. Studies have shown that Alzheimer’s polygenic risk score also predicts correlates of Alzheimer’s disease risk, including memory decline (15) and structural perturbations in the hippocampus (2, 16–18), in adults without dementia. Measures of cognitive function in childhood and early adulthood predict later risk (4, 19). Nevertheless, results on the association between APOE-ε4 allele and family history of Alzheimer’s disease and brain structure in children and adolescents have been inconsistent (20–22).
Despite these interesting clues, no study has investigated associations among aggregate genetic risk factors for Alzheimer’s disease, cognition measures, and brain volumes in children and adolescents. This approach may increase our understanding of Alzheimer’s disease pathogenesis and risk, potentially enabling the development of preventive measures, early diagnosis, and better treatments targeting initial signs of the disease.
In this study, we investigated the associations of Alzheimer’s polygenic risk score in childhood and adolescence with cognitive performance, reading and writing abilities, executive function, and hippocampal volume in two Brazilian samples: a discovery sample from Porto Alegre (N=364) and a replication sample from São Paulo (N=352). Data were collected at the two sites using the same data collection procedures and cognitive tasks and similar scanners and MRI acquisition protocols. In addition, we searched for evidence of comparable associations in a third sample of Canadian adolescents (N=1,029), assessed with distinct cognitive tasks and MRI acquisition protocol. We hypothesized that children with a higher polygenic risk score perform worse on memory recall, reading, and writing tests and have lower hippocampal volumes.
Method
Brazilian Samples
Participants were children and adolescents 6 to 14 years of age from the Brazilian High Risk Study for Psychiatric Disorders (23). Participants were recruited from schools in two cities in Brazil: Porto Alegre (arbitrarily defined as the Brazilian discovery sample before any data analysis) and São Paulo (defined as the Brazilian replication sample). Participants were selected at both sites by a screening phase encompassing 9,937 children and followed by a selection of two subgroups: a high-risk subgroup for psychiatric disorders composed of 1,554 participants with psychiatric symptoms and high family loading of symptoms, and a random-selection subgroup with 958 individuals (23).
From this pool of 2,512 participants, a total of 716 (364 in Porto Alegre and 352 in São Paulo) underwent genotyping. Within this subsample, all children (N=716) performed reading and writing tests, 668 performed the Rey-Osterrieth Complex Figure Test (ROCFT), 677 performed executive function tests, and 670 underwent T1-weighted structural MRI.
Parents of the participants and literate participants provided written consent, and verbal agreement was obtained from illiterate participants. The Ethics Committee of the University of São Paulo approved the study. Detailed information regarding the selection of participants has been published elsewhere (23).
Genotyping.
EDTA tubes were used to collect whole blood. Genomic DNA was isolated using Gentra Puregene kits (Qiagen). Genotyping was performed using the HumanOmniExpress V1 (Illumina). Single-nucleotide polymorphisms (SNPs) with a minor allele frequency <1%, locus missingness >10%, or Hardy-Weinberg equilibrium significance <0.000001 were excluded, as were individuals with genotype missingness >10% and an estimation of identity by descent >0.12. We performed genotype imputation in the Michigan Imputation Server (https://imputationserver.sph.umich.edu), using the 1000G phase 1, version 3, and the prephasing algorithm SHAPEIT2.
Polygenic risk score.
Alzheimer’s polygenic risk score was calculated with the PRSice software package (24), using as a training sample the summary statistics of the International Genomics of Alzheimer’s Project (IGAP) (http://web.pasteur-lille.fr/en/recherche/u744/igap/igap_download.php), and as a target sample our imputed genotype. P value–informed clumping was performed retaining the SNP with smallest p value within a 250-kb window and excluding SNPs that were in linkage disequilibrium (r2>0.1). For the main analyses, we selected, a priori, a p threshold of 0.01, which contained 5,116 independent SNPs in the training and target samples, because it had been previously used in other studies using Alzheimer’s polygenic risk score (2). Supplemental analyses investigating other thresholds using PRSice are provided in the data supplement that accompanies the online edition of this article (section 2.2).
Nondeclarative memory.
Nondeclarative memory was assessed using the ROCFT. The ROCFT is widely used to assess nondeclarative visuospatial memory as well as attention, coordination, and organizational skills. Impairment indicated on the ROCFT has been linked to Alzheimer’s diagnosis in adults (25) and to family-genetic risk for Alzheimer’s disease in children (19).
The ROCFT involves three steps: in the first (copy), individuals are asked to draw the figure while looking at the ROCFT stimulus card; in the second (immediate recall) and third (delayed recall), participants are asked to draw the figure from memory, with immediate recall tested at 3 minutes and delayed recall at 30 minutes. Performance was assessed by trained raters blind to all other data, via the Quantitative Scoring System (26). With this scoring system, each of 18 items concerning location and shape accuracy was assigned a score from 0 to 2.
ROCFT scores were calculated using the mean percentage of retained items for each item (i.e., recall score/copy score), excluding items in which the recall score outperformed the copy score, for both immediate and delayed recall. We also excluded participants who drew less than 50% of the ROCFT items in the copy task (N=6). (Further information regarding the reliability and validity of the ROCFT and the score methods used in this study is provided in the online data supplement, section 1.) Dependent variables were factor scores for each task after regressing out age trends by saving studentized residuals.
Reading and writing abilities.
Participants’ reading and writing abilities were quantified using the Brazilian version of the Academic Performance Test (27). In the reading task, participants were asked to read aloud 70 words, and in the writing task, they were asked to write 34 dictated items. Dependent variables were factor scores for each task after regressing out age trends by saving studentized residuals.
Executive function.
Participants performed six tasks for assessment of working memory, inhibitory control, and time processing, which loaded on a single higher-order executive function factor. The second-order model provided excellent fit to the data, as described elsewhere (28). The dependent variables were factor scores for the second-order model after regressing out age trends by saving studentized residuals. (Further information about the assessment of executive function is provided in the data supplement, section 4.)
Hippocampal volumes.
T1-weighted structural MR images were acquired using 1.5-T GE Signa HDx (Brazilian replication sample) and Signa HD (Brazilian discovery sample) scanners with the following parameters: TR=10.916 ms, TE=4.2 ms, slice thickness=1.2 mm, flip angle=15°, matrix size=256×192, FOV=245 mm, max=156 slices). Imaging acquisitions were repeated whenever subjects moved during the procedure until optimal quality was obtained.
To estimate hippocampal volumes, we used the Multiple Automatically Generated Templates for different brains (MAGeT) algorithm (29), a newly developed multi-atlas segmentation method. Further information regarding the MAGeT algorithm can be found elsewhere (30, 31). Hippocampal volumes were then adjusted by total intracranial volume by retaining studentized residuals in a linear regression model estimated using FreeSurfer, version 5.3. A total of 12 participants were excluded in quality control of the MAGeT algorithm.
Canadian Sample
Participants in the Canadian sample were 1,024 adolescents (mean age, 15 years [SD=0.8]; 52% were female) from 481 families, recruited from high schools in Quebec. All adolescents had European (French) ancestry and at least one sibling 12–18 years old. Further information about this sample can be found elsewhere (32).
Briefly, nondeclarative memory was assessed using the dot location subtest and the stories subtest of the Children’s Memory Scale (33). In the dot location subtest, participants were asked to place eight chips on a blank grid in locations previously shown. This procedure was repeated five times, forming a total raw score. After 40 minutes, participants were asked to recall the location of the first pattern (long delay). Both long delay and total raw scores were then divided by learning score in order to reduce bias from task misunderstanding. In the stories subtest, participants were asked to listen to a story read aloud by the examiner and to repeat it aloud promptly (immediate recall) and again after 20–30 minutes (delayed recall). They also performed a delayed recognition task, in which they were asked yes-or-no questions about the stories. All dependent variables were factor scores for the second-order model after regressing out age trends by saving studentized residuals.
Genotyping was performed using the Illumina Human610-Quad BeadChip and HumanOmniExpress BeadChip. Alzheimer’s polygenic risk scores were calculated in the same way as described for the Brazilian samples. Again, we selected a p threshold of 0.01 a priori. Neuroimaging was conducted on a Phillips 1.0-T superconducting magnet, with T1-weighted images acquired using the following parameters: three-dimensional radiofrequency spoiled gradient echo scan with 140–160 slices, 1-mm isotropic resolution, TR=25 ms, TE=5 ms, and flip angle=30°. Hippocampal volumes were also estimated using the MAGeT algorithm. Further information can be found elsewhere (34).
Statistical Analysis
The analyses for the Brazilian samples were tested using mixed-effect models to investigate the associations between Alzheimer’s polygenic risk score (independent variable; p<0.01 threshold selected a priori) and immediate and delayed recall factor scores, reading and writing factor scores, executive function factor scores, and hippocampal volumes (dependent variables). In the ROCFT, reading, and writing analyses, we included tester and participant’s school as random variables. Given that our main samples were from an admixed Brazilian population, four of the principal components of GWAS were used as covariates in all mixed-model analyses, as suggested by previous studies (35). All analyses were adjusted by sampling weights, which adjust for our high risk selection procedure (28). Dependent (e.g., memory performance) and independent variables (i.e., Alzheimer’s polygenic risk score) were standardized to reflect standardized regression coefficients (β) and to facilitate interpretation.
After the main analysis, we conducted two sets of exploratory analyses and four sets of sensitivity analyses. Exploratory analyses included an analysis to assess the best p threshold for each dependent variable using regression analysis in PRSice and an analysis assessing whether associations vary as a function of the level of polygenic risk score (i.e., whether participants with high scores would have disproportionately affected phenotypes).
Sensitivity analyses for significant results included an analysis specific to Caucasian subjects (N=428); an analysis investigating associations between outcomes and APOE alleles (using the following imputation algorithm: reference Panel 1000G, phase 3, version 5; phasing: Eagle, version 2.3); an analysis investigating associations between outcomes and each SNP composing the polygenic risk score composite to investigate the biological meaning of the results; and a generalizability test to investigate whether the results would replicate in a completely different sample of Canadian adolescents (N=1,029). All analyses in the Canadian sample used siblinghood as a random variable and used four principal components of the GWAS as covariates.
Results
Participants
In comparison with the original Brazilian sample (N=2,512), individuals who underwent genotyping had a similar mean age (original sample: mean=124.6 months, SD=23.05; genotyped sample: mean=121.2 months, SD=22.31), sex distribution (original sample: 46.7% female; genotyped sample: 45.9% female), and IQ (original sample: mean=100.49, SD=16.07; genotyped sample: mean=101.14, SD=16.27).
Compared with participants in the Brazilian replication sample, the Brazilian discovery sample had a higher mean age, a higher percentage of Caucasians, a lower mean family income, and a higher mean Alzheimer’s polygenic risk score (Table 1).
| Characteristic | Brazilian Discovery Sample (N=364) | Brazilian Replication Sample (N=352) | ||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| Age (years) | 10.26 | 1.98 | 9.95 | 1.73 |
| IQ | 100.39 | 16.56 | 101.86 | 15.97 |
| Family income (USD) | 890.40 | 693.30 | 1223.54 | 734.10 |
| Alzheimer’s disease polygenic risk score | 0.00156 | 0.00047 | 0.00170 | 0.00041 |
| N | % | N | % | |
| Female | 154 | 46.4 | 138 | 39.9 |
| Ethnicity | ||||
| Caucasian | 221 | 66.7 | 187 | 54.0 |
| Black | 47 | 14.2 | 21 | 6.1 |
| Multiracial | 60 | 18.2 | 136 | 39.3 |
| Indigenous | 3 | 0.9 | 1 | 0.3 |
| Asian | 0 | 0.0 | 1 | 0.3 |
TABLE 1. Characteristics of the Brazilian Discovery and Replication Samples in a Study of Polygenic Risk Score for Alzheimer’s Disease
Nondeclarative Memory
Analyses showed that for the Brazilian discovery sample, a one-unit increase in z-score for Alzheimer’s polygenic risk score predicted a 0.185 z-score decrement in immediate recall performance (p=0.012) and a 0.282 decrement in delayed recall performance (p<0.0001). These findings were replicated for both analyses in the Brazilian replication sample (immediate recall: β=−0.259, p=0.003; delayed recall: β=−0.232; p=0.008). Univariate analyses also revealed significant and replicable associations for both immediate recall (Brazilian discovery sample: β=−0.148, p=0.002; Brazilian replication sample: β=−0.197, p=0.001) and delayed recall (Brazilian discovery sample: β=−0.211, p<0.001; Brazilian replication sample: β=−0.215, p<0.001). Results are depicted in Figure 1.
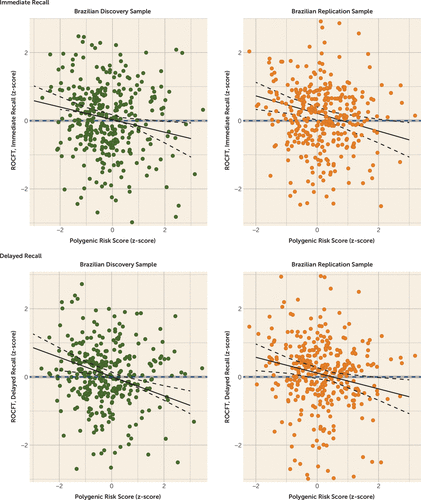
FIGURE 1. Association Between Immediate and Delayed Recall Z-Scores and Polygenic Risk Score for Alzheimer’s Disease in the Brazilian Discovery and Replication Samplesa
a ROCFT=Rey-Osterrieth Complex Figure Test.
Exploratory PRSice analyses examining multiple thresholds for the Brazilian total sample revealed maximum explanation of the phenotype variability at threshold p<0.0119 (NSNPs=5,845) for immediate recall (p=0.008) and at threshold p<0.02025 (NSNPs=8,880) for delayed recall (p=0.002) (see section 2.2 in the online data supplement). Associations had no differences in effect size across several levels of Alzheimer’s polygenic risk score in quantile regressions (see section 2.3 in the data supplement).
Reading and Writing Abilities
Analyses showed no association between Alzheimer’s polygenic risk score and either reading (β=0.039, p=0.600) or writing scores (β=0.066, p=0.407) in the Brazilian discovery sample. Of note, there were significant and replicable associations in univariate analyses for reading (discovery sample: β=−0.199, p<0.001; replication sample: β=−0.158, p=0.009) and writing (discovery sample: β=−0.161, p=0.003; replication sample: β=−0.165, p=0.007), which may reflect uncontrolled confounding.
Exploratory PRSice analyses evaluating multiple thresholds for the Brazilian total sample showed maximum explanation of the phenotype variability at threshold p<0.026 (NSNPs=10,740) for reading (p=0.013) and writing (p=0.033) (see section 2.2 in the data supplement). Associations had no differences in effect size across several levels of Alzheimer’s polygenic risk score in quantile regressions (see section 2.3 in the data supplement).
Executive Function
Analyses showed no association between Alzheimer’s polygenic risk score and executive function (β=−0.002; p=0.979) for the Brazilian discovery sample. Exploratory PRSice analyses evaluating multiple thresholds also failed to find significant associations for other thresholds (see section 2.2 in the data supplement). Associations had no differences in effect size across several levels of Alzheimer’s polygenic risk score in quantile regressions (see section 2.3 in the data supplement). Of note, there were significant and replicable associations in univariate analyses for executive function (Brazilian discovery sample: β=−0.139, p<0.001; Brazilian replication sample: β=−0.099, p=0.042), which may reflect uncontrolled confounding.
Hippocampal Volumes
Analyses revealed a positive association between Alzheimer’s polygenic risk score and right hippocampal volume (β=0.150, p=0.045) for the Brazilian discovery sample, which was not replicated in the Brazilian replication sample (β=−0.002, p=0.982). No association was found for left hippocampal volume in either sample (discovery sample: β=0.085, p=0.243; replication sample: β=0.077, p=0.376).
Exploratory PRSice analyses evaluating multiple thresholds for the Brazilian total sample showed significant associations for other thresholds, which reached maximum explanation of the phenotype at risk-score threshold p<0.132 for left hippocampal volume (NSNPs=38,878; p=0.044), with several other thresholds also explaining statistically significant levels of variance. For the right hippocampal volume, a model fit reached maximum explanation at threshold p<0.1185 (NSNPs=35,863; p=0.009) (see section 2.2 in the data supplement).
Quantile regressions suggest that both left and right hippocampal volumes exhibited associations with Alzheimer’s polygenic risk score in participants who had high polygenic risk scores (see section 2.3 in the data supplement). We found negative associations between hippocampal volume and polygenic risk score in the Brazilian total sample when risk score was above the 82nd percentile in quantile regressions for the left hemisphere (β=−0.156, 95% CI=−0.265, −0.013) and above the 75th percentile for the right hemisphere (β=−0.114, 95% CI=−0.192, −0.0004) (Figure 2). A joint test comparing the equality of the slopes also showed significant differences from the median for both the 82nd percentile (F=7.004, p=0.008) and the 75th percentile (F=5.193, p=0.023) for left and right hippocampal volumes, respectively, which revealed differences in the slope estimations for median and high polygenic risk scores. We did not find associations between hippocampal volumes and cognitive measures (data available on request).
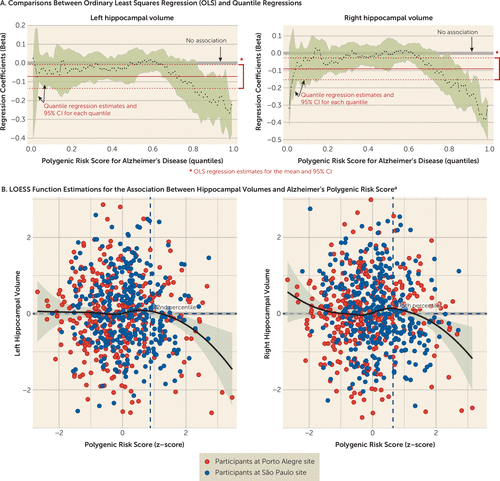
FIGURE 2. Association Between Left and Right Hippocampal Volumes and Polygenic Risk Score for Alzheimer’s Disease Using Quantile Regression and LOESS Function Estimations for the Brazilian Total Sample
a In panel B, estimations were performed with the LOESS function and tested statistically using quantile regressions. Dotted lines indicate the 82nd and the 75th percentile of Alzheimer’s polygenic risk score for left and right hippocampal volumes, respectively.
The level of Alzheimer’s polygenic risk score that was associated with lower hippocampal volume differed between the Brazilian discovery and replication samples. In the discovery sample, significant associations emerged at the 96th percentile for the left hippocampus (β=−0.301, 95% CI=−0.434, −0.087) and above the 91st percentile for the right hippocampus (β=−0.319, 95% CI=−0.468, −0.072). In the replication sample, however, significant findings emerged above the 63rd percentile for the left hippocampus (β=−0.127, 95% CI=−0.211, −0.005) and above the 59th percentile for the right hippocampus (β=−0.111, 95% CI=−0.219, −0.020).
These analyses were extended by examining associations with hippocampal subregions. The right CA4 and dentate gyrus were associated with Alzheimer’s polygenic risk score at the main threshold in both the Brazilian discovery and replication samples. However, this analysis was performed post hoc and not defined a priori (see section 2.4 in the data supplement). Moreover, no other subregions exhibited replicable associations with the Alzheimer’s polygenic risk score.
Sensitivity Analyses
Brazilian Caucasian subsample.
For the Brazilian Caucasian subsample (N=428), we found significant associations between Alzheimer’s polygenic risk score and both immediate (β=−0.155, p=0.006) and delayed recall (β=−0.212, p=0.0001). We found no association between polygenic risk score and hippocampal volumes in quantile regressions. Results were similar for the Brazilian non-Caucasian subsample (see section 2.1 in the data supplement).
Associations with APOE alleles.
We found no association between APOE alleles and immediate (F=1.919, p=0.091) or delayed (F=0.984, p=0.427) recall, or either right (F=1.152, p=0.333) or left (F=1.235, p=0.293) hippocampal volume. Moreover, associations between Alzheimer’s polygenic risk score and memory performance among participants with the more frequent APOE genotype group (3/3 alleles; N=469) were also significant for immediate (β=−0.181, p=0.006) and delayed (β=−0.253, p<0.001) recall. Thus, our results are unlikely to be driven solely by the APOE ε4 allele.
Assessing the role of specific SNPs composing the Alzheimer’s polygenic risk score and the biological significance of the findings.
We found no significant associations between each individual SNP from the Alzheimer’s polygenic risk score (p<0.01 threshold) and memory performance or hippocampal volumes after correction for multiple comparisons (p<9.7×10−6). This suggests that aggregate weighted risk arises from the SNPs included in the score rather than from specific associations with one or another SNP (see section 5 in the data supplement). The MAGMA software package, using the IGAP summary statistics for SNPs’ p values and Reactome as background, found several enriched pathways, including immunoregulatory interactions between lymphoid and nonlymphoid cells (R-HSA-198933), VLDL assembly (R-HSA-8866423), and Netrin-1 signaling (R-HSA-373752).
Canadian sample.
In the dot location subtest, associations fell short of significance for total score (β=−0.053, p=0.097) and long delay score (β=−0.059, p=0.065). In the stories subtest, no association was found for immediate recall (β=0.002, p=0.947), delayed recall (β=0.040, p=0.220), and stories recognition (β=0.008, p=0.781). We detected no associations between Alzheimer’s polygenic risk score and either right (β=0.047, p=0.127) or left (β=0.025, p=0.464) hippocampal volume. No significant results were found in quantile regressions.
Discussion
Polygenic risk score for Alzheimer’s disease was associated with lower scores in both immediate and delayed recall in nondeclarative memory tasks in children and adolescents from two independent samples from Brazil. For the genetic risk threshold defined a priori, no replicable associations were found for verbal abilities, executive function, and hippocampal volumes in the Brazilian sample analyses. Nevertheless, we found suggestive evidence of associations with reading, writing, and hippocampal volumes using other risk thresholds for Alzheimer’s polygenic risk score. Moreover, we found that the association between polygenic risk score and hippocampal volumes varies with level of polygenic risk score. In the Canadian sample, associations that fell short of significance were observed for total score and long delay score.
Our findings are in agreement with previous studies that showed an association of Alzheimer’s polygenic risk score with memory decline (15) and lower hippocampal volume (2, 16, 29) in adults. Our findings extend previous evidence showing that these associations also occur in children, long before the onset of the disease. As in previous studies, these findings suggest that even classical neurodegenerative diseases such as Alzheimer’s disease may have neurodevelopmental roots (4), like other chronic disorders of adults (36). The possibility of a neurodevelopmental origin creates opportunities for research on Alzheimer’s disease prevention, risk detection, and pathogenesis.
Interestingly, associations with memory performance were shown for every level of Alzheimer’s polygenic risk score in a linear way. However, similar to some studies investigating young adults (29), but unlike others (2, 16), we found no replicable associations with hippocampal volumes for the threshold selected a priori. Nevertheless, we did detect associations when using alternative thresholds and when considering individuals with a particularly high Alzheimer’s polygenic risk score. This may reflect a nonlinear association with genetic risk for hippocampal volumes, consistent with previous findings (37). In addition, in the present study, associations were not accounted for by the presence of the APOE-ε4 allele or any other particular SNP allele but rather appeared to arise from aggregate genetic risk. The lack of associations with APOE-ε4 may suggest that the impact of this allele on the predisposition to Alzheimer’s disease may be clinically apparent only later in life. It also raises the hypothesis that the effects from SNPs associated with Alzheimer’s disease in early life may influence vulnerability to APOE effects later in life.
Our significant findings were not generalizable to a third sample of Canadian adolescents, assessed with a distinct protocol. The lack of associations found for the Canadian sample may reflect multiple factors, including the possibility that our significant results represent a type I error. Nevertheless, replication in two independent Brazilian samples studied with identical assessment protocols decreases this possibility and suggests the need to consider alternative explanations, including the use of distinct Alzheimer’s polygenic risk scores for each sample, unique interactions with genetic background, and specificity to one or another cognitive test. Other work supports some of these possibilities. Given that African Americans have an increased risk of developing Alzheimer’s disease (38), the Brazilian admixed population could have more power to detect influences of Alzheimer’s polygenic risk score in cognitive functions, as compared with the Canadian sample. This hypothesis is also supported by the association found between Alzheimer’s polygenic risk score quintiles and ethnicity (Caucasian versus non-Caucasian) (χ2=141.023, p<0.001). Furthermore, previous studies did not show differences in memory performance using the Children’s Memory Scale in children with genetic predisposition to Alzheimer’s disease (39), as opposed to the ROCFT (19), and therefore the lack of replication in the Canadian sample may represent some specificity to the memory task used in each sample. It is also important to emphasize that these tasks are not specific to one domain, which may contribute to the differences found in these analyses.
Our study has some limitations that need to be addressed. First, the Alzheimer’s polygenic risk score was generated based on a Caucasian sample, and the SNPs associated with Alzheimer’s disease could be different for other ethnicities and races present in our admixed Brazilian samples. However, similar results were found for a Brazilian Caucasian subsample in our sensitivity analyses, which decreases the likelihood of false positives due to population stratification. Second, the density of our SNP array (∼250K SNPs) did not cover several SNPs analyzed in previous Alzheimer’s polygenic risk score studies. Nevertheless, this score was able to successfully predict both cognition and brain volume in individuals with high genetic risk.
Our findings suggest that Alzheimer’s polygenic risk score may influence developmental processes underlying nondeclarative visuoconstructive memory and hippocampal volume in early life, expanding the understanding of the influences of SNPs associated with Alzheimer’s disease before the diagnosis of the disease and providing further mechanisms for identifying individuals at higher risk of developing Alzheimer’s disease. Further research is needed to replicate these findings in other samples and to advance our understanding of mechanisms linking genetic risk for Alzheimer’s disease and the development of cognitive functions.
1 : Alzheimer’s disease: a clinical and basic science review. Med Student Res J 2014; 4:24–33Google Scholar
2 : Polygenic risk of Alzheimer disease is associated with early- and late-life processes. Neurology 2016; 87:481–488Crossref, Medline, Google Scholar
3 : Reminiscence therapy in dementia: a review. Maturitas 2012; 72:203–205Crossref, Medline, Google Scholar
4 : Linguistic ability in early life and the neuropathology of Alzheimer’s disease and cerebrovascular disease: findings from the Nun Study. Ann N Y Acad Sci 2000; 903:34–38Crossref, Medline, Google Scholar
5 : Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry 2006; 63:168–174Crossref, Medline, Google Scholar
6 : How heritable is Alzheimer’s disease late in life? Findings from Swedish twins. Ann Neurol 2004; 55:180–185Crossref, Medline, Google Scholar
7 : Whole-genome analyses of whole-brain data: working within an expanded search space. Nat Neurosci 2014; 17:791–800Crossref, Medline, Google Scholar
8 : Genome-wide association studies in Alzheimer’s disease. Hum Mol Genet 2009; 18(R2):R137–R145Crossref, Medline, Google Scholar
9 : Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet 2009; 41:1088–1093Crossref, Medline, Google Scholar
10 : Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 2013; 45:1452–1458Crossref, Medline, Google Scholar
11 : Effects of multiple genetic loci on age at onset in late-onset Alzheimer disease: a genome-wide association study. JAMA Neurol 2014; 71:1394–1404Crossref, Medline, Google Scholar
12 : Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 2010; 303:1832–1840Crossref, Medline, Google Scholar
13 : Genome-wide association studies in Alzheimer disease. Arch Neurol 2008; 65:329–334Crossref, Medline, Google Scholar
14 : Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain 2015; 138:3673–3684Crossref, Medline, Google Scholar
15 : Using an Alzheimer disease polygenic risk score to predict memory decline in black and white Americans over 14 years of follow-up. Alzheimer Dis Assoc Disord 2016; 30:195–202Crossref, Medline, Google Scholar
16 : Multimodal brain imaging reveals structural differences in Alzheimer’s disease polygenic risk carriers: a study in healthy young adults. Biol Psychiatry 2017; 81:154–161Crossref, Medline, Google Scholar
17 : The association between a polygenic Alzheimer score and cortical thickness in clinically normal subjects. Cereb Cortex 2012; 22:2653–2661Crossref, Medline, Google Scholar
18 : An Alzheimer’s disease genetic risk score predicts longitudinal thinning of hippocampal complex subregions in healthy older adults. eNeuro 2016; 3:ENEURO.0098-16.2016Crossref, Google Scholar
19 : Decreased cognition in children with risk factors for Alzheimer’s disease. Biol Psychiatry 2008; 64:904–906Crossref, Medline, Google Scholar
20 : Alzheimer gene APOE ε4 linked to brain development in infants. JAMA 2014; 311:298–299Crossref, Medline, Google Scholar
21 : Cortical morphology in children and adolescents with different apolipoprotein E gene polymorphisms: an observational study. Lancet Neurol 2007; 6:494–500Crossref, Medline, Google Scholar
22 : No differences in hippocampal volume between carriers and non-carriers of the ApoE ε4 and ε2 alleles in young healthy adolescents. J Alzheimers Dis 2014; 40:37–43Crossref, Medline, Google Scholar
23 : High risk cohort study for psychiatric disorders in childhood: rationale, design, methods, and preliminary results. Int J Methods Psychiatr Res 2015; 24:58–73Crossref, Medline, Google Scholar
24 : PRSice: Polygenic Risk Score software. Bioinformatics 2015; 31:1466–1468Crossref, Medline, Google Scholar
25 : Performance on the Rey-Osterrieth Complex Figure Test in Alzheimer disease and vascular dementia. Neuropsychiatry Neuropsychol Behav Neurol 1999; 12:95–101Medline, Google Scholar
26 : Rey Complex Figure Test and Recognition Trial Professional Manual. Lutz, Fla, Psychological Assessment Resources, 1995Google Scholar
27 : TDE: Teste de Desempenho Escolar: Manual para Aplicação e Interpretação. São Paulo, Casa Psicólogo, 1994Google Scholar
28 : A general psychopathology factor (P factor) in children: structural model analysis and external validation through familial risk and child global executive function. J Abnorm Psychol 2017; 126:137–148Crossref, Medline, Google Scholar
29 : The effect of increased genetic risk for Alzheimer’s disease on hippocampal and amygdala volume. Neurobiol Aging 2016; 40:68–77Crossref, Medline, Google Scholar
30 : Multi-atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates. Neuroimage 2014; 101:494–512Crossref, Medline, Google Scholar
31 : A novel in vivo atlas of human hippocampal subfields using high-resolution 3 T magnetic resonance imaging. Neuroimage 2013; 74:254–265Crossref, Medline, Google Scholar
32 : Cohort profile: the Saguenay Youth Study (SYS). Int J Epidemiol 2017; 46:e19Medline, Google Scholar
33 : CMS: Children’s Memory Scale: Manual. San Antonio, Tex, Psychological Corp, 1997Google Scholar
34 : Estimating volumes of the pituitary gland from T1-weighted magnetic-resonance images: effects of age, puberty, testosterone, and estradiol. Neuroimage 2014; 94:216–221Crossref, Medline, Google Scholar
35 : Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38:904–909Crossref, Medline, Google Scholar
36 : Fetal origins of mental health: the developmental origins of health and disease hypothesis. Am J Psychiatry 2017; 174:319–328Link, Google Scholar
37 : Toward a dynamic biomarker model in Alzheimer’s disease. J Alzheimers Dis 2012; 30:91–100Crossref, Medline, Google Scholar
38 : A meta-analysis of Alzheimer’s disease incidence and prevalence comparing African-Americans and Caucasians. J Alzheimers Dis 2016; 50:71–76Crossref, Medline, Google Scholar
39 : Apolipoprotein E4 and sex affect neurobehavioral performance in primary school children. Pediatr Res 2010; 67:293–299Crossref, Medline, Google Scholar



