Common Genetic Variation and Antidepressant Efficacy in Major Depressive Disorder: A Meta-Analysis of Three Genome-Wide Pharmacogenetic Studies
Abstract
Objective
Indirect evidence suggests that common genetic variation contributes to individual differences in antidepressant efficacy among individuals with major depressive disorder, but previous studies may have been underpowered to detect these effects.
Method
A meta-analysis was performed on data from three genome-wide pharmacogenetic studies (the Genome-Based Therapeutic Drugs for Depression [GENDEP] project, the Munich Antidepressant Response Signature [MARS] project, and the Sequenced Treatment Alternatives to Relieve Depression [STAR*D] study), which included 2,256 individuals of Northern European descent with major depressive disorder, and antidepressant treatment outcomes were prospectively collected. After imputation, 1.2 million single-nucleotide polymorphisms were tested, capturing common variation for association with symptomatic improvement and remission after up to 12 weeks of antidepressant treatment.
Results
No individual association met a genome-wide threshold for statistical significance in the primary analyses. A polygenic score derived from a meta-analysis of GENDEP and MARS participants accounted for up to approximately 1.2% of the variance in outcomes in STAR*D, suggesting a weakly concordant signal distributed over many polymorphisms. An analysis restricted to 1,354 individuals treated with citalopram (STAR*D) or escitalopram (GENDEP) identified an intergenic region on chromosome 5 associated with early improvement after 2 weeks of treatment.
Conclusions
Despite increased statistical power accorded by meta-analysis, the authors identified no reliable predictors of antidepressant treatment outcome, although they did identify modest, direct evidence that common genetic variation contributes to individual differences in antidepressant response.
Antidepressant medications have repeatedly demonstrated greater efficacy than placebo in the treatment of major depressive disorder (1, 2). However, individual patients vary widely in antidepressant treatment response, and only about one-third of patients achieve symptomatic remission with an initial treatment (3). Several indirect lines of evidence suggest that genetic variation may contribute to this variability. These include observations of familiality of response to antidepressants in relatively small family studies (4–6), as well as animal studies indicating quantitative trait loci associated with antidepressant-related behavioral phenotypes (7, 8).
However, to date, no consistently replicated findings have emerged from genetic association studies of antidepressant efficacy. One possible explanation is that if antidepressant response is a polygenic phenotype associated with common variation, individual studies have been underpowered to detect all but the largest effects. In other heritable phenotypes, such as type 2 diabetes, coronary artery disease, rheumatoid arthritis, and inflammatory bowel disease, the combination of studies in meta-analyses has led to success in identifying association with common variation, even when individual studies have been unsuccessful in identifying such association (9–11). The same has held true for neuropsychiatric disorders, including schizophrenia and bipolar disorder (12, 13).
In an effort to identify single-nucleotide polymorphisms (SNPs) associated with antidepressant response, we combined results from the three genome-wide pharmacogenetic studies of antidepressant efficacy in major depression published to date: the Genome-Based Therapeutic Drugs for Depression (GENDEP) project (14, 15), the Munich Antidepressant Response Signature (MARS) project (16, 17), and the Sequenced Treatment Alternatives to Relieve Depression (STAR*D) study (18, 19). We hypothesized that a meta-analysis would identify robust associations that are more likely to replicate in independent data sets. To pursue the competing goals of maximum power and minimum heterogeneity, we performed two analyses: a broader analysis that included all patients in order to reveal non-treatment-specific pharmacogenetic associations and a narrower analysis restricted to patients treated with either of two selective serotonin reuptake inhibitors (SSRIs) (citalopram or escitalopram).
Method
Samples
The GENDEP project is a 12-week multicenter part-randomized open-label pharmacogenetic trial with two active treatment arms: protocol-guided escitalopram (10–30 mg/day) and the tricyclic antidepressant nortriptyline (50–150 mg/day), which is a norepinephrine-reuptake inhibitor. Treatment was provided for 12 weeks on an outpatient basis (14). Inclusion criteria were a diagnosis of moderate to severe unipolar depression according to ICD-10/DSM-IV criteria, as determined by the Schedules for Clinical Assessment in Neuropsychiatry interview (20), age 18 to 75 years, and Caucasian ancestry, defined as having four grandparents of white European origin. The primary outcome measure was the Montgomery-Åsberg Depression Rating Scale (MADRS) (21), administered weekly by psychiatrists and psychologists with high reliability. Of the 811 recruited adult patients, 706 (87%) passed phenotype and genotype quality control and were included in genome-wide analyses (15). GENDEP was approved by ethics boards of the participating centers, and written informed consent was obtained from all participants. Demographic and clinical characteristics of the study participants are summarized in Table 1.
| Characteristic | GENDEP | MARS | STAR*D (Level 1) | |||
|---|---|---|---|---|---|---|
| N | % | N | % | N | % | |
| Participants included in meta-analysis | 672 | 604 | 980 | |||
| Participants treated with selective serotonin reuptake inhibitors (SSRIs) | 374 | 56 | NAb | 980 | 100 | |
| Female | 429 | 64 | 326 | 54 | 580 | 59 |
| Valid outcomes for at least 4 weeks of treatment | 597 | 89 | 532 | 88 | 943 | 96 |
| Remission by week 12 | 270 | 46 | 253 | 47 | 330 | 35 |
| Partial response at week 2 | 256 | 38 | 400 | 67 | 268 | 27 |
| Mean | SD | Mean | SD | Mean | SD | |
| Age (years) | 41.9 | 11.7 | 48.4 | 14.0 | 43.6 | 13.2 |
| Baseline 17-item Hamilton Depression Rating Scale score | 22.4 | 4.7 | 25.1 | 5.6 | 21.3 | 5.1 |
| Percentage change on primary measure over first 2 weeks of treatment | 21.1 | 22.3 | 35.0 | 26.4 | 23.9 | 23.6 |
| Percentage change on primary measure over 12 weeks of treatment | 55.5 | 30.7 | 63.7 | 27.8 | 56.5 | 28.1 |
The MARS project is a prospective naturalistic study of a representative sample of adult inpatients admitted to hospitals in southern Germany for depression (16). Inclusion criteria were a diagnosis of a major depressive episode (first-episode major depressive disorder, recurrent major depressive disorder, or bipolar disorder) based on DSM-IV criteria and a clinical interview by trained psychiatrists; age 18 to 75 years; and Caucasian ancestry. Treatment was selected naturalistically by clinicians and included flexible dosage of antidepressants and augmenting agents (16). The primary outcome measure was the Hamilton Depression Rating Scale (HAM-D), administered weekly by trained psychiatrists and psychologists. Of the 842 participants recruited by 2008, 339 were included in a previously reported genome-wide pharmacogenetic study (17), and additional samples from this cohort have been genotyped since then, resulting in 604 (72%) samples from patients with unipolar depression available for the present meta-analysis. MARS was approved by the ethics committee of Ludwig Maximilians University, and all participants provided written consent after the study protocol and potential risks were explained. Demographic and clinical characteristics of the study participants are summarized in Table 1.
The STAR*D study is a pragmatic trial of protocol-guided antidepressant treatment for outpatients with major depression (19). The study included 4,041 treatment-seeking adult outpatients, recruited in 18 primary care and 23 psychiatric clinical sites across the United States. Inclusion criteria were a diagnosis of nonpsychotic unipolar major depressive disorder diagnosed by a clinician and confirmed with a checklist of DSM-IV criteria; age 18 to 75 years; and a minimum score of 14 on the HAM-D. The present meta-analysis uses data from the first treatment step, which included protocol-guided citalopram (20–60 mg/day) (22). Depression severity in STAR*D was rated every 2 weeks using the clinician-rated and self-report versions of the 16-item Quick Inventory for Depressive Symptomatology (QIDS) (23). The primary outcome measure was the 17-item HAM-D, administered by trained independent evaluators at study entry and at the end of each treatment step (19). However, since data from QIDS were available for more participants and this assessment tool was found to be closely equivalent to HAM-D, most STAR*D reports rely on it primarily (22, 24). Genetic material was collected from 1,948 (48%) participants; of whom 1,491 (37% of the original STAR*D sample, including 980 of white/European ancestry) passed quality control and were included in previously reported genome-wide analyses (18). The study was approved by institutional ethics review boards at all centers. Written consent was obtained from all participants after the procedures and any associated risks were explained. STAR*D genotype and phenotype data are available through the National Institute of Mental Health Human Genetic Initiative (https://www.nimhgenetics.org/). Demographic and clinical characteristics of the study participants are summarized in Table 1.
Common Inclusion Criteria
Although the inclusion and exclusion criteria of the three component studies overlapped, there were several differences. To minimize heterogeneity, we imposed three common inclusion criteria for our meta-analysis.
First, homogeneous ethnicity was required for each component analysis to minimize the risk of confounding due to population stratification. White European/Caucasian ethnicity was an inclusion criterion in the GENDEP and MARS studies. Of the STAR*D genetic sample, 72% of participants were non-Hispanic white/European Americans, 16% were black/African Americans, and 12% were Latino/Hispanic. Thus, the STAR*D sample included in our meta-analysis was limited to 980 white/European Americans (72% of those who were otherwise eligible).
Second, unipolar major depression (i.e., the absence of a personal history of hypomanic, manic, or mixed episodes) was a requirement in the GENDEP and STAR*D studies. In the MARS study, 11% of participants had bipolar disorder. Since response to antidepressants may differ between unipolar and bipolar depression (25), our meta-analysis was restricted to individuals with unipolar depression. As a result, 604 (89% of those who were otherwise eligible) MARS participants were included in our analysis.
Third, a minimum depression severity score of 14 on the 17-item HAM-D, corresponding to recommendations for a quantitative definition of moderate depression (26, 27), was an inclusion criterion in the MARS and STAR*D studies but not in the GENDEP study. Since specific antidepressant response is associated with severity (1), only individuals with a score ≥14 at baseline were included in our meta-analysis. As a result, 672 (95% of those who were otherwise eligible) GENDEP participants were included in our analysis.
Demographic and clinical characteristics of the GENDEP, MARS, and STAR*D participants that passed our common inclusion criteria are summarized in Table 1.
Phenotype Definition
The therapeutic response to antidepressants evolves over a number of weeks, and the optimal definition of outcome has been subject to debate (28–30). Traditionally, outcome of antidepressant treatment in clinical trials has been defined as a categorical (yes/no) variable, based on a predefined cutoff value on a rating scale at study exit (e.g., a HAM-D score ≤7 defines remission) or a cutoff value on the relative improvement expressed as a proportion of severity score reduction from study entry (e.g., an improvement of ≥50% defines response). Categorical measures are easily presented and understood but use only part of the available information and are strongly influenced by study duration, dropouts, and initial severity (31, 32). Continuous measures of change (e.g., percentage of change from baseline) capture more information and can be adjusted for baseline variables and the effects of dropouts or discontinuation before planned study exit, but they are more difficult to present and translate into clinical decisions. Since investigators differ in their preferences and the three component studies differed in the use of either continuous (14, 30) or categorical (17, 18) outcome measures, our meta-analysis plan specified two primary outcome measures: one continuous and one categorical.
The primary continuous outcome measure was percentage improvement on the clinician-rated depression scale in each study over up to 12 weeks of treatment, corrected for age, sex, and recruitment center. The MADRS was used in the GENDEP study, the HAM-D in the MARS study, and the 16-item clinician-rated QIDS in the STAR*D study. In case of dropout before week 12, the missing data were estimated from earlier measurements, based on the best linear unbiased predictor from mixed-effects models as previously described and recommended (14, 15, 33). All individuals with at least one valid postbaseline measurement of depression severity were included in this analysis.
The primary categorical outcome measure was remission, defined as a HAM-D score ≤7 on the last available measurement of depression severity or an equivalent score on the MADRS (a score of 10) or on the clinician-rated QIDS (a score of 5), with no imputation of missing data. Since the potential to achieve remission depends on the duration of active treatment, only individuals with valid data on depression severity after at least 4 weeks of antidepressant treatment were included.
In addition, two secondary outcomes of interest were defined to evaluate genetic contribution to the early changes over the first 2 weeks of antidepressant treatment. The secondary continuous outcome was percentage change in depression severity over the first 2 weeks of treatment, corrected for age, sex, and recruitment center. The secondary categorical outcome was early partial response, defined as a 25% improvement on the HAM-D (or equivalent rating on the MADRS or the clinician-rated QIDS) after the first 2 weeks of antidepressant treatment (17). All outcomes of interest and analytic methods were defined prior to initiating meta-analysis.
Genotyping and Imputation
In the three component studies, DNA was extracted from blood or lymphoblastoid cell lines and genotyped on arrays measuring one-half million or more SNPs that tag the majority of common variants in the human genome. The GENDEP and MARS samples were genotyped using the Illumina Human610-Quad BeadChip (Illumina, Inc., San Diego). STAR*D samples were genotyped using the Affymetrix Human Mapping 500K Array and the Genome-Wide Human SNP Array 5.0 (Affymetrix, Santa Clara, Calif.). Quality control to exclude SNPs with low call rates, admixture, cryptic relatedness, and abnormal heterozygosity rates, as well as SNPs from contaminated or degraded samples or samples with low genotyping success, was carried out separately in each study as previously reported (15, 17, 18). Data on additional markers were imputed using BEAGLE 3.3 (34) and with HapMap phase-3 CEU (Centre d’Etude du Polymorphisme Humain from Utah population) as the reference data set, resulting in a common set of 1.2 million markers.
The analytic plan specified that any SNPs significant at a genome-wide significance level that relied on inaccurately imputed data (i.e., an imputation information score <0.8) in one or more cohorts would be regenotyped. TaqMan was used in the GENDEP study, while the MARS and STAR*D studies used a Sequenom MALDI-TOF (matrix-assisted laser desorption/ionization time-of-flight) mass-spectrometer platform with iPlex technology (Sequenom, San Diego).
Statistical Analysis and Power
In each study, the effects of genotypes on treatment outcomes were tested using linear regression for continuous outcomes and logistic regression for categorical outcomes, applied using PLINK (35). To account for uncertainty of imputation, these analyses were performed for dosage data, with estimated probability of each genotype. To minimize the risk of confounding through population stratification, significant principal components or dimensions describing the structure of each data set were included as covariates. We also controlled for age, sex, and recruitment center, either by adjusting outcome prior to analyses (for continuous outcomes) or by inclusion of these factors as covariates (for categorical outcomes), since they are more likely to be confounders than intermediate phenotypes on the pathway between a genetic disposition and response to treatment. Factors such as personality and comorbidity were not included in the analyses, since they are more likely to be intermediate phenotypes with a strong genetic contribution and may represent mediators rather than moderators of association.
We carried out a fixed-effects meta-analysis using the weighted Z method in METAL (36), which represents the standard approach in genome-wide studies and allows comparison with other reports. To test whether the assumption of homogeneity of effect underlying fixed-effects meta-analyses was met, we also carried out heterogeneity tests (Cochrane’s Q statistic and the I2 heterogeneity index) and, for completeness, random-effects meta-analyses using PLINK (35).
Two meta-analyses were performed. First, an overall analysis of data from 2,256 participants tested the hypothesis that common genetic variants contribute to the outcome of treatment with various antidepressant drugs across the three component studies. Second, we performed a drug-specific meta-analysis of the escitalopram-treated GENDEP participants (N=374) and the citalopram-treated STAR*D participants (N=980) to test the hypothesis that common genetic variants predict outcome of treatment with SSRIs. A genome-wide significance threshold was set at the generally accepted p value of 5×10−8 (37). A suggestive significance and reporting threshold was set at a p value of 5×10−6, which is two orders of magnitude below the genome-wide significance level and approximately corresponds to a level at which one association per genome-wide analysis is expected by chance (37). Results of associations with a p value <1×10−4 are reported in the data supplement that accompanies the online edition of this article.
Assuming consistent effect across studies (38), our meta-analysis had a power of 86% to detect an additive genetic effect explaining 2% of the variance in the continuous outcome at the genome-wide significance level (p<5×10−8) and 86% power to detect an outcome explaining 1.5% of the variance at the suggestive level of significance (p<5×10−6) in the entire sample. Assuming a minor allele frequency of 0.25, the test of additive genetic effect on the categorical outcome of remission had 81% power to detect an odds ratio of 1.35 at the genome-wide significance level (p<5×10−8) and 84% power to detect an odds ratio of 1.3 at the suggestive level of significance (p<5×10−6). The analysis restricted to SSRI-treated participants had a power of ≥80% to detect an additive genetic effect explaining 3.5% of the variance or a SNP (minor allele frequency=0.25) associated with an odds ratio of 1.5 at the genome-wide significance level (p<5×10−8).
Both the overall meta-analysis and the meta-analysis restricted to SSRI-treated patients had a power of 99% to detect, at a genome-wide level of significance, clinically significant associations (39). However, multiple weak pharmacogenetic associations may remain undetected. Therefore, in addition to single variant analyses, polygenic scores were constructed to test the joint effect of multiple weak associations across the genome. Specifically, for the primary outcomes, polygenic scores were constructed based on a meta-analysis of the two smaller studies (GENDEP and MARS) with the number of risk alleles weighted by strength of association after removing SNPs with low minor allele frequency (<0.02), excluding the major histocompatibility complex region, and pruning for linkage disequilibrium (R2<0.25) so that SNPs that share more than 80% of the variance were not included, leaving 117,000 independent SNPs for potential inclusion in polygenic scores (13). Polygenic scores were calculated as a weighted (by effect size) sum of risk alleles across markers associated at a p-value threshold. Ten scores were calculated based on progressive p-value thresholds (<0.0001, 0.001, 0.01, 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, and 1.0). The resulting scores were tested as predictors of improvement and remission in STAR*D using linear and logistic regression, respectively. The proportion of variance explained was estimated as R2 in linear regression and as the Nagelkerke pseudo R2 in logistic regression. This means that the two estimates are not directly comparable.
Results
Meta-Analyses
Primary outcomes: improvement and remission with up to 12 weeks of antidepressant treatment in the entire sample
First, we performed a meta-analysis of 12-week outcomes in the entire sample of 2,256 patients with major depressive disorder (Table 1). Quantile-quantile plots (see Figure S1 in the online data supplement) and lambda scores between 0.99 and 1.02 revealed no departures from uniform distributions of p values across approximately 1.2 million genotyped and imputed markers.
The results of the fixed-effects meta-analysis are summarized in Figure 1 and Table S1 in the online data supplement. The relatively rare imputed SNP rs17651119 (minor allele frequency=0.014) located in an intronic region of the myosin X (MYO10) gene at 5p15.1 was associated with percentage improvement in the initial analysis (p=1.78×10−8), but follow-up genotyping yielded a reduced association (beta=–0.24; p=0.045) because of an absence of association in STAR*D. Suggestive associations (p<5×10−6) with percent improvement were found for four independent SNPs (rs2546057, rs12410462, rs17634917, and rs264272; p≤3.87×10−6). For the outcome measure of remission, four independent SNPs met the suggestive threshold (rs9601248, rs2125000, rs17710780, and rs9466930; p≤4.45×10−6).

a Data are from the Genome-Based Therapeutic Drugs for Depression (GENDEP) project, the Munich Antidepressant Response Signature (MARS) project, and the Sequenced Treatment Alternatives to Relieve Depression (STAR*D) study. Remission was measured using the Hamilton Depression Rating Scale. The y-axis plots indicate p values for associations on the negative logarithmic scale (–log10[p values]). Gene symbols indicate the gene on which the associated single-nucleotide polymorphism (SNP) (p≤5×10−6) is located, or, if the gene symbol is in parentheses, the nearest gene up to 100 kb away from the associated SNP. An imputed SNP located in an intronic region of the myosin X (MYO10) gene at 5p15.1 achieved a genome-wide effect, which could not be validated in confirmatory follow-up genotyping.
Polygenic scores constructed based on a meta-analysis of improvement and remission in the GENDEP and MARS studies significantly predicted improvement and remission in STAR*D (Figure 2). For remission, the scores with the 10 progressive p-value thresholds included 46; 388; 3,469; 15,122; 27,876; 50,449; 70,463; 88,195; 104,156; and 156,601 SNPs.
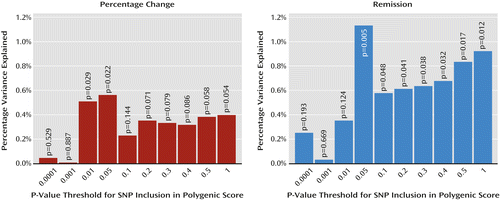
a GENDEP=Genome-Based Therapeutic Drugs for Depression; MARS=Munich Antidepressant Response Signature; STAR*D=Sequenced Treatment Alternatives to Relieve Depression. The x-axis indicates the meta-analysis p-value threshold for single-nucleotide polymorphism inclusion. The y-axis indicates the percentage of variance explained in STAR*D.
Secondary outcomes: early improvement and partial response after 2 weeks of treatment in the entire sample
No genome-wide significant association was found for the 2-week outcomes (Figure 3 and Table S2 in the online data supplement). For percentage improvement at 2 weeks, three suggestive associations were identified (rs7174755, rs10065906, and rs12513663; p≤2.47×10−6). For early partial response (25% improvement at 2 weeks), three such associations were also noted (rs10065906, rs10174573, and rs166040; p≤2.47×10−6). One of these, rs10065906, located in an intergenic region at 5q33.3, revealed suggestive associations with both secondary outcomes (early improvement: p=1.99×10−6; early partial response: p=5.29×10−8).
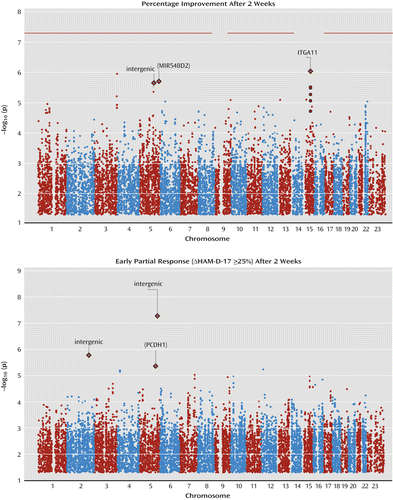
a GENDEP=Genome-Based Therapeutic Drugs for Depression; MARS=Munich Antidepressant Response Signature; STAR*D=Sequenced Treatment Alternatives to Relieve Depression. Early partial response was defined as a 25% improvement on the Hamilton Depression Rating Scale. The y-axis plots p values for associations on the negative logarithmic scale (–log10[p values]). Gene symbols indicate the gene on which the associated single-nucleotide polymorphism (SNP) (p≤5×10−6) is located, or, if the gene symbol is in parentheses, the nearest gene up to 100 kb away from the associated SNP.
12-week outcomes with SSRIs
All of the 980 participants from the STAR*D study and 374 (out of 672) participants from the GENDEP study were treated with an SSRI (citalopram or escitalopram). In our meta-analysis of these 1,354 individuals, we searched for polymorphisms associated with the efficacy of SSRIs, using the two primary and two secondary phenotypes. The same analytic methods and quality-control criteria used for the entire sample were applied here. Quantile-quantile plots and lambda scores between 1.01 and 1.04 revealed near-uniform distributions of p values, suggesting no effects of population stratification (see Figure S2 in the online data supplement).
For the primary continuous outcome of relative improvement over up to 12 weeks of treatment, no SNP was associated at the genome-wide level of significance (see Table S3 and Figure S3 in the online data supplement). Five suggestive associations were detected (rs17538444, rs1034394, rs264272, rs6598266, and rs398426; p≤4.51×10−6), including an intronic SNP (rs17538444; p=4.17×10−7) in the ENOX1 gene, encoding an electron transporter and oxidase.
For the primary categorical outcome of remission after up to 12 weeks of treatment, no SNP predicted outcome at a genome-wide level of significance (see Table S3 and Figure S3 in the online data supplement). Three suggestive associations (rs1525293, rs364477, and rs8012941; p≤4.48×10−6) included an intronic SNP (rs8012941; p=4.48×10−7) in the KCNH5 gene, which encodes a voltage-gated potassium channel.
2-week outcomes with SSRIs
For the secondary categorical outcome of early partial response at 2 weeks, there were no genome-wide significant associations and two markers associated at a suggestive level of significance (rs6799788, rs10065906; p≤1.69×10−6; see Table S4 and Figure S4 in the online data supplement).
Discussion
This meta-analysis integrates the majority of currently available genome-wide association data on antidepressant response in individuals with major depressive disorder, and, to our knowledge, represents the largest combined pharmacogenetic sample for any psychotropic medication. Notwithstanding substantial differences in the design of the three primary studies analyzed, it was possible to establish common inclusion criteria, outcome measures, imputation procedures, and clinical analytical methods to minimize heterogeneity.
Taken together, the three cohorts yielded statistical power to allow detection of individual variants explaining between 1% and 2% of variance in antidepressant response. In primary and secondary analyses, no single variant met the criteria for genome-wide significance. Confirmatory genotyping of rs17651119, located in an intronic region of the myosin X (MYO10) gene at 5p15.1, did not support an initial genome-wide association signal in imputed data.
The failure to identify individual common variants of large effect is consistent with other genome-wide association studies of complex diseases. Typically, meta-analyses of 5,000 or more case and comparison subjects have been required to begin to reliably detect the more modest associations anticipated in such disorders (9–12). The primary rationale for the present meta-analysis was the success in detecting associations with more extreme treatment-response phenotypes in smaller cohorts outside of psychiatry. For example, a modestly sized cohort was sufficient to identify association with a variant contributing risk for myopathy in statin-treated patients (40). The lack of strong associations in the present meta-analysis suggests that unlike dramatic drug toxicity phenotypes, antidepressant response will likely be moderated by numerous modest genetic effects.
A methodology examining the composite effects of a large number of variants of more modest effect, even when individual variants have not been identified, has been described and validated in disorders such as schizophrenia (13). We applied this approach to generate polygenic scores based on the meta-analyzed MARS and GENDEP cohorts and examined the variance accounted for in the third independent cohort, STAR*D. The polygenic risk score accounts for between 0.5% and 1% of variance. While previous investigations have examined familiality of antidepressant response (4–6), as far as we are aware, our results represent the first direct demonstration of common genetic risk influencing antidepressant response, suggesting that strategies using larger cohorts and more homogeneous or extreme phenotypes may succeed in identifying specific variants.
One encouraging preliminary result comes from our analysis restricted to SSRI-treated individuals drawn from the STAR*D cohort and escitalopram-treated individuals in GENDEP. This analysis identified a variant associated with early SSRI response (within the first 2 weeks of treatment) at a threshold considered to be genome-wide significant, although it would not survive further correction for the number of phenotypes examined. This variant tags a linkage disequilibrium block of approximately 200 kb, including 15 SNPs (r2>0.60) showing suggestive associations with the same phenotype. This region appears to be in an intergenic region on chromosome 5, between 31 and 175 kb from a cluster of predicted genes (e.g., LOC643401) but with no evidence of transcription. As with most such reported findings, if it can be replicated, further investigation will be required to understand its functional significance.
Another limitation is the heterogeneity inherent in combining data from trials that differ in design, recruitment strategy, and treatment selection. We used common inclusion criteria to make the samples of the three studies more comparable on the most important characteristics. While this does not completely eliminate between-study heterogeneity, pharmacogenetic effects that are narrowly specific to more homogeneous populations are unlikely to be applicable in practice. We elected to pool across treatment groups in order to maximize power to detect drug effects, based on the assumption that genetic moderators of response are similar across classes of antidepressants. However, this hypothesis is untested, and the heterogeneity of treatment reduced the power to detect drug-specific pharmacogenetic effects. We therefore performed a second meta-analysis that excluded the MARS cohort and was restricted to individuals treated with citalopram or escitalopram, two antidepressants with nearly identical pharmacological properties (45). STAR*D and GENDEP, outpatient studies of first-line antidepressants in nonpsychotic patients, have proven sufficiently homogeneous to allow robust replication of clinical associations (46).
Overall, our results suggest the complex genetic architecture of antidepressant response and the need for larger cohorts of systematically treated and prospectively observed subjects. Results from genome-wide studies of other phenotypes indicate that this approach can succeed when larger sample sizes are achieved. Our report may provide a foundation for such efforts in antidepressant response.
1 : Antidepressant drug effects and depression severity: a patient-level meta-analysis. JAMA 2010; 303:47–53Crossref, Medline, Google Scholar
2 : Efficacy of antidepressants: a re-analysis and re-interpretation of the Kirsch data. Int J Neuropsychopharmacol 2011; 14:405–412Crossref, Medline, Google Scholar
3 : Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: a STAR*D report. Am J Psychiatry 2006; 163:1905–1917Link, Google Scholar
4 : Effect of antidepressives and genetic factors. Arzneimittelforschung 1964; 14(Suppl):496–500Medline, Google Scholar
5 : Familial concordance of fluvoxamine response as a tool for differentiating mood disorder pedigrees. J Psychiatr Res 1998; 32:255–259Crossref, Medline, Google Scholar
6 : Pharmacogenetic response to antidepressants in a multicase family with affective disorder. Biol Psychiatry 1994; 36:467–471Crossref, Medline, Google Scholar
7 : Strain-dependent antidepressant-like effects of citalopram in the mouse tail suspension test. Psychopharmacology (Berl) 2005; 183:257–264Crossref, Medline, Google Scholar
8 : Pharmacogenomic evaluation of the antidepressant citalopram in the mouse tail suspension test. Neuropsychopharmacology 2006; 31:2433–2442Crossref, Medline, Google Scholar
9 : Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet 2011; 43:246–252Crossref, Medline, Google Scholar
10 : TCF7L2 is reproducibly associated with type 2 diabetes in various ethnic groups: a global meta-analysis. J Mol Med (Berl) 2007; 85:777–782Crossref, Medline, Google Scholar
11 : The genome-wide association study—a new era for common polygenic disorders. J Cardiovasc Transl Res 2010; 3:173–182Crossref, Medline, Google Scholar
12 : Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet 2008; 40:1056–1058Crossref, Medline, Google Scholar
13 : Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460:748–752Crossref, Medline, Google Scholar
14 : Differential efficacy of escitalopram and nortriptyline on dimensional measures of depression. Br J Psychiatry 2009; 194:252–259Crossref, Medline, Google Scholar
15 : Genome-wide pharmacogenetics of antidepressant response in the GENDEP project. Am J Psychiatry 2010; 167:555–564Link, Google Scholar
16 : Clinical characteristics and treatment outcome in a representative sample of depressed inpatients - findings from the Munich Antidepressant Response Signature (MARS) project. J Psychiatr Res 2009; 43:215–229Crossref, Medline, Google Scholar
17 : A genomewide association study points to multiple loci that predict antidepressant drug treatment outcome in depression. Arch Gen Psychiatry 2009; 66:966–975Crossref, Medline, Google Scholar
18 : A genomewide association study of citalopram response in major depressive disorder. Biol Psychiatry 2010; 67:133–138Crossref, Medline, Google Scholar
19 : Sequenced Treatment Alternatives to Relieve Depression (STAR*D): rationale and design. Control Clin Trials 2004; 25:119–142Crossref, Medline, Google Scholar
20 : Diagnosis and Clinical Measurement in Psychiatry: A Reference Manual for SCAN. Geneva, World Health Organization, 1998Crossref, Google Scholar
21 : Measuring depression: comparison and integration of three scales in the GENDEP study. Psychol Med 2008; 38:289–300Crossref, Medline, Google Scholar
22 : Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: implications for clinical practice. Am J Psychiatry 2006; 163:28–40Link, Google Scholar
23 : An evaluation of the Quick Inventory of Depressive Symptomatology and the Hamilton Rating Scale for Depression: a Sequenced Treatment Alternatives to Relieve Depression trial report. Biol Psychiatry 2006; 59:493–501Crossref, Medline, Google Scholar
24 : Selecting among second-step antidepressant medication monotherapies: predictive value of clinical, demographic, or first-step treatment features. Arch Gen Psychiatry 2008; 65:870–880Crossref, Medline, Google Scholar
25 : Antidepressants for the acute treatment of bipolar depression: a systematic review and meta-analysis. J Clin Psychiatry 2011; 72:156–167Crossref, Medline, Google Scholar
26 : A comparison of depression rating scales. Br J Psychiatry 1982; 141:45–49Crossref, Medline, Google Scholar
27 : Hamilton Depression Rating Scale. Extracted from Regular and Change Versions of the Schedule for Affective Disorders and Schizophrenia. Arch Gen Psychiatry 1981; 38:98–103Crossref, Medline, Google Scholar
28 : Trajectories of change in depression severity during treatment with antidepressants. Psychol Med 2010; 40:1367–1377Crossref, Medline, Google Scholar
29 : The definition and operational criteria for treatment outcome of major depressive disorder. A review of the current research literature. Arch Gen Psychiatry 1991; 48:796–800Crossref, Medline, Google Scholar
30 : A genomewide association study of citalopram response in major depressive disorder-a psychometric approach. Biol Psychiatry 2010; 68:e25–e27Crossref, Medline, Google Scholar
31 : Dichotomizing continuous predictors in multiple regression: a bad idea. Stat Med 2006; 25:127–141Crossref, Medline, Google Scholar
32 : Power considerations when a continuous outcome variable is dichotomized. J Biopharm Stat 1998; 8:337–352Crossref, Medline, Google Scholar
33 : A systematic method for estimating individual responses to treatment with antipsychotics in CATIE. Schizophr Res 2009; 107:13–21Crossref, Medline, Google Scholar
34 : A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 2009; 84:210–223Crossref, Medline, Google Scholar
35 : PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81:559–575Crossref, Medline, Google Scholar
36 : METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26:2190–2191Crossref, Medline, Google Scholar
37 : Estimation of significance thresholds for genomewide association scans. Genet Epidemiol 2008; 32:227–234Crossref, Medline, Google Scholar
38 : Meta-analysis of genome-wide association studies: no efficiency gain in using individual participant data. Genet Epidemiol 2010; 34:60–66Medline, Google Scholar
39 : Biomarkers predicting treatment outcome in depression: what is clinically significant? Pharmacogenomics 2012; 13:233–240Crossref, Medline, Google Scholar
40 : The SLCO1B1*5 genetic variant is associated with statin-induced side effects. J Am Coll Cardiol 2009; 54:1609–1616Crossref, Medline, Google Scholar
41 : Placebo response in studies of major depression: variable, substantial, and growing. JAMA 2002; 287:1840–1847Crossref, Medline, Google Scholar
42 : A new population-enrichment strategy to improve efficiency of placebo-controlled clinical trials of antidepressant drugs. Clin Pharmacol Ther 2010; 88:634–642Crossref, Medline, Google Scholar
43 : Estimating drug effects in the presence of placebo response: causal inference using growth mixture modeling. Stat Med 2009; 28:3363–3385Crossref, Medline, Google Scholar
44 : Translating biomarkers to clinical practice. Mol Psychiatry 2011; 16:1076–1087Crossref, Medline, Google Scholar
45 : The pharmacological effect of citalopram residues in the (S)-(+)-enantiomer. J Neural Transm 1992; 88:157–160Crossref, Medline, Google Scholar
46 : Depression symptom dimensions as predictors of antidepressant treatment outcome: replicable evidence for interest-activity symptoms. Psychol Med 2012; 42:967–980Crossref, Medline, Google Scholar



