From Rare Copy Number Variants to Biological Processes in ADHD
Abstract
Objective:
Attention deficit hyperactivity disorder (ADHD) is a highly heritable psychiatric disorder. The objective of this study was to define ADHD-associated candidate genes and their associated molecular modules and biological themes, based on the analysis of rare genetic variants.
Methods:
The authors combined data from 11 published copy number variation studies in 6,176 individuals with ADHD and 25,026 control subjects and prioritized genes by applying an integrative strategy based on criteria including recurrence in individuals with ADHD, absence in control subjects, complete coverage in copy number gains, and presence in the minimal region common to overlapping copy number variants (CNVs), as well as on protein-protein interactions and information from cross-species genotype-phenotype annotation.
Results:
The authors localized 2,241 eligible genes in the 1,532 reported CNVs, of which they classified 432 as high-priority ADHD candidate genes. The high-priority ADHD candidate genes were significantly coexpressed in the brain. A network of 66 genes was supported by ADHD-relevant phenotypes in the cross-species database. Four significantly interconnected protein modules were found among the high-priority ADHD genes. A total of 26 genes were observed across all applied bioinformatic methods. Lookup in the latest genome-wide association study for ADHD showed that among those 26 genes, POLR3C and RBFOX1 were also supported by common genetic variants.
Conclusions:
Integration of a stringent filtering procedure in CNV studies with suitable bioinformatics approaches can identify ADHD candidate genes at increased levels of credibility. The authors’ analytic pipeline provides additional insight into the molecular mechanisms underlying ADHD and allows prioritization of genes for functional validation in validated model organisms.
Attention deficit hyperactivity disorder (ADHD) is one of the most common neuropsychiatric disorders, with a prevalence of 5%−6% in children (1). The disorder persists into adulthood in a significant proportion of affected individuals, resulting in a prevalence of 2.5%−4.9% in adults (2). The clinical symptoms of ADHD include age-inappropriate inattention, hyperactivity, and impulsivity (3). Twin and adoption studies have estimated a heritability of 76% for ADHD (3).
Identification of the genes implicated in ADHD and their molecular functions offers opportunities to understand the neurobiological mechanisms leading to ADHD and facilitates the development of diagnostic tools and new treatments. However, despite the high heritability, identification of ADHD risk genes has been difficult, mainly because of ADHD’s complex genetic architecture (2, 4, 5). To date, mainly genetic variants that frequently occur in the population have been investigated for their role in ADHD, either through studies of candidate genes or hypothesis-free genome-wide association studies (GWASs) (3, 6). A recent GWAS meta-analysis identified the first 12 loci harboring ADHD risk variants (7). Another type of GWAS has focused on the association of rare copy number variants (CNVs) with ADHD. Such CNV GWASs have largely concentrated on rare events (primarily) observed in individuals diagnosed with ADHD. We analyzed the 11 studies published to date that have detected rare CNVs in ADHD case subjects (8–18). Those CNV GWASs implicated more than 2,200 candidate genes in ADHD, although most have investigated rather limited sample sizes, and most of the CNVs were detected only in single patients. Based on the average mutation rate between individuals in the general population, single-patient rare findings have a high chance of being false positives; it is thus important to concentrate on repeatedly occurring copy number events (19).
Data integration from various sources is an important strategy for moving from genes to biologically meaningful modules. Examples for this come from publications on ADHD and related disorders (20, 21). A study on autism spectrum disorder (20) showed how data integration enabled the identification of highly conserved gene clusters that improve our understanding of neuropsychiatric disorders. Similarly, a recent study (21) found a significant overlap of ADHD case CNVs with targets of the fragile X mental retardation protein (FMRP), a gene cluster involved in neurodevelopmental disorder risk. In many cases, data integration currently takes place only across data modalities derived from studies in humans. This neglects the wealth of phenotypic information that can be derived from model organisms such as monkey, rat, mouse, zebrafish, and fruit fly (6).
In this study, we surveyed and integrated data on CNVs associated with ADHD from existing publications, aiming to define robustly ADHD-associated genes, molecular modules, and biological themes underlying this disorder. We combined data from the 11 published ADHD CNV studies and applied an integrative strategy using redundancy criteria, data on protein-protein interactions, and employing information from cross-species genotype-phenotype annotation to prioritize candidate genes (22, 23). We classified 432 high-priority ADHD candidate genes, supported by coexpression, cross-species phenotype, and protein interaction information, with 26 genes highlighted across all approaches. Integration with data on common genetic variants showed that among these 26 genes, POLR3C and RBFOX1 were significantly associated with ADHD in the largest single-nucleotide polymorphism (SNP)–based GWAS meta-analysis to date.
Methods
Identification of Genes Affected by ADHD CNVs
Coordinates of rare CNVs occurring in individuals diagnosed with ADHD were retrieved from the 11 studies published thus far (8–18). Discovery samples of 6,176 ADHD case subjects and 25,026 control subjects in total provided the basis for the analyzed studies (for study characteristics, see Table S1 in the online supplement). Independence of the samples was ensured by evaluating the cohorts of the included studies. Duplicates of samples that overlapped between studies (occurring between Williams et al. [9] and Stergiakouli et al. [11]) were removed. Coordinates of 1,532 CNVs were retrieved for our analysis. These were mapped to the same reference human genome (hg19), using University of California Santa Cruz (UCSC) Lift Genome Annotations; the minimal ratio of bases that must remap was set to 0.95 (https://genome.ucsc.edu/cgi-bin/hgLiftOver). The CNV coordinates were used to retrieve RefGene information from the UCSC MySQL database, using a Structured Query Language (SQL) query (see the online supplement) (genome-mysql.cse.ucsc.edu). Information retrieved included the overlap with coding sequence, transcriptional direction, the total gene and CNV size in base pairs, exact gene start/end position, and percentage of gene coding sequence represented by the CNV (see Table S2, spreadsheet tab 1, in the online supplement).
Selection of Genes Recurrently Affected in ADHD CNVs
Transcript variants and biotypes were extracted for each CNV through a batch National Center for Biotechnology Information nucleotide query (for biotypes, see Table S2, spreadsheet tab 1, in the online supplement). For gene copy number losses, we included all genes that were entirely deleted or partially truncated. We also annotated whether the N- or C-terminal region of transcripts was affected (see Table S2, spreadsheet tab 1). For gene copy number gains, we considered transcripts for which both a 2-kb promoter and the coding region were entirely duplicated. For overlapping CNVs, the minimal chromosomal region of overlap was identified to narrow down the putative region involved in ADHD. Only mRNA-coding genes affected by CNVs in at least two case subjects with ADHD were selected for subsequent analyses (high-priority catalog), given interpretability and the possibility of performing cluster and protein-protein interaction analyses.
Coexpression Network Analysis
The BrainSpan developmental transcriptome data set (RNA-Seq Gencode, version 10) was used to investigate the overrepresentation of coexpressed genes in our high-priority gene set across all brain regions and developmental time points (embryo to adult) relative to the rest of the genome (24). The expression coefficients for each mRNA-coding gene at all time points and in all brain regions in the BrainSpan data set were concatenated. The coexpression correlation score was calculated for each gene pair. Gene pairs with a correlation score >0.3 were assigned to a coexpression network, each node representing a single gene and each connection representing the correlation score. The sum of the correlation scores for the investigated gene set and for 10,000 random gene sets of the same size and coding sequence length was calculated. An enrichment score was calculated by dividing the sum of the correlation scores per gene set by the mean of the 10,000 random gene sets. The p values were calculated by comparing how many of the 10,000 correlation scores of the random gene sets were equal to or higher than those of the investigated gene set.
Integrated Cross-Species Phenotype and Protein-Protein Interaction Network
We used the Monarch Initiative cross-species phenotype database to retrieve genes associated with an ADHD-related phenotype (25). We defined core phenotypes by selecting terms based on attention deficit, hyperactivity, and impulsivity (see Table S3 in the online supplement). The genes connected to the cross-species core phenotypes of ADHD were subsequently superimposed onto interspecies Biological General Repository for Interaction Datasets (BioGRID) protein interaction data. The interaction plot was visualized with Cytoscape, version 3.4.0 (26).
Identification of Enriched-Protein Interaction Modules
Networks of physical interactions in the gene set were assessed with the Disease Association Protein-Protein Link Evaluator (DAPPLE, version 0.17), a widely used algorithm described in detail elsewhere (27, 28). We used InWeb data with the following parameter settings: number of permutations: 1,000 with adaptive permutation function; plot: true; seed file: genes. A set of 432 high-priority genes formed the input for this analysis. The modules were visualized with Cytoscape.
Gene-Based Association Analyses of Prioritized Modules With ADHD Risk
We used data from the recent meta-analysis of GWASs of 20,183 patients with ADHD and 35,191 control subjects performed by the Psychiatric Genomics Consortium (PGC) ADHD working group and the Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH) (7). In addition, data from the genome-wide meta-analysis across eight psychiatric disorders (N=438,997) were used (both are publicly available at https://www.med.unc.edu/pgc/results-and-downloads/). (Details on the samples and quality control can be found in the Supplementary Methods section of the online supplement and in references 7 and 29.)
Gene-based association analyses were performed using the Multimarker Analysis of GenoMic Annotation (MAGMA) software package, version 1.05 (for details, see the Supplementary Methods section of the online supplement) (30). In the analyses using data from the cross-disorder meta-analysis, one gene (of the 26 prioritized genes) was not included, SEPT5, because there were no SNPs included in this genetic region in the filtered summary statistics. Genes were considered gene-wide significant if they reached the Bonferroni correction threshold adjusted for the number of genes tested (p<0.05/26).
Brain Gene Expression
GENE2FUNC, a core process of FUMA (Functional Mapping and Annotation of Genome-wide Association Studies; http://fuma.ctglab.nl [31]), was employed to analyze gene expression patterns. For this analysis, the set of 26 genes most consistently identified was used as input, including prioritized genes (Table 1). Gene expression heat maps were constructed employing GTEx, version 8 (54 tissue types) and BrainSpan RNA-seq data across 29 different ages. The average of normalized expression per label (zero means across samples) was displayed on the corresponding heat maps. Expression values are TPM (transcripts per million) for GTEx and RPKM (reads per kilobase million) for the BrainSpan data set. Heat maps display normalized expression value (zero mean normalization of log2 transformed expression), and darker red means higher relative expression of that gene in each label, compared with a darker blue color in the same label.
| Gene Symbol | Full Gene Name | Gene Function | Biological Themes |
|---|---|---|---|
| COPA | Coatomer Protein Complex Subunit Alpha | Protein transport between the endoplasmic reticulum and Golgi compartments | |
| CPSF2 | Cleavage and Polyadenylation Specific Factor 2 | Pre-mRNA 3′-end formation | mRNA metabolism |
| CSMD1 | CUB And Sushi Multiple Domains 1 | Unknown | |
| DMRTB1 | DMRT Like Family B With Proline Rich C-Terminal 1 | Transcription factor | Transcription |
| MAPK1 | Mitogen-Activated Protein Kinase 1 | Wide range of cellular processes, e.g., proliferation, differentiation, transcription regulation, and development | |
| MSH3 | MutS Homolog 3 | DNA mismatch repair | |
| MYC | MYC Proto-Oncogene, BHLH Transcription Factor | Transcription factor | Transcription |
| NCSTN | Nicastrin | Type I transmembrane glycoprotein | |
| NDUFA5 | NADH:Ubiquinone Oxidoreductase Subunit A5 | Subunit of complex I of the mitochondrial respiratory chain | Mitochondria |
| NDUFB1 | NADH:Ubiquinone Oxidoreductase Subunit B1 | Subunit of complex I of the mitochondrial respiratory chain | Mitochondria |
| PARK2 | Parkin RBR E3 Ubiquitin Protein Ligase | Component of a multiprotein E3 ubiquitin ligase complex, degradation of defective mitochondria | Mitochondria |
| PDCD6IP | Programmed Cell Death 6 Interacting Protein | Functions within the ESCRT pathway | |
| PEA15 | Phosphoprotein Enriched in Astrocytes 15 | Negative regulator of apoptosis | |
| PLOD3 | Procollagen-Lysine,2-Oxoglutarate 5-Dioxygenase 3 | Homodimeric enzyme in the rough endoplasmic reticulum | |
| POLR1A | RNA Polymerase I Subunit A | Subunit of the RNA polymerase I complex | mRNA metabolism |
| POLR2B | RNA Polymerase II Subunit B | Subunit of the RNA polymerase II complex | mRNA metabolism |
| POLR3C | RNA Polymerase III Subunit C | Subunit of the RNA polymerase III complex | mRNA metabolism |
| PPM1F | Protein Phosphatase, Mg2+/Mn2+Dependent 1F | Ser/Thr protein phosphatase | |
| RBFOX1 | RNA Binding Protein, Fox-1 Homolog 1 | Regulates tissue-specific alternative splicing | |
| SEPT5 | Septin 5 | Regulation of cytoskeletal organization and SNARE complexes | Cytoskeleton |
| TBL1XR1 | Transducin Beta Like 1 X-Linked Receptor 1 | F-box-like protein involved in the recruitment of the ubiquitin/19S proteasome complex to nuclear receptor–regulated transcription units | Transcription |
| TTC27 | Tetratricopeptide Repeat Domain 27 | Unknown | |
| TUBA3C | Tubulin Alpha 3c | Major component of microtubules | Cytoskeleton |
| TUBGCP2 | Tubulin Gamma Complex Associated Protein 2 | Microtubule nucleation at the centrosome | Cytoskeleton |
| WASL | Wiskott-Aldrich Syndrome Like | Regulates actin polymerization | Cytoskeleton |
| WWOX | WW Domain-Containing Oxidoreductase | WW domain-containing oxidoreductase |
TABLE 1. The 26 genes consistently identified across all bioinformatics approaches employed in the study
Results
Identification of Genes Located in ADHD-Associated CNVs and Definition of High-Priority Gene Set
We extracted data from the 11 studies reporting rare CNVs in a total discovery sample of 6,176 ADHD case subjects and 25,026 control subjects (for study characteristics, see Table S1 in the online supplement). Coordinates of 1,532 CNVs were retrieved, containing 2,241 mRNA-coding candidate genes.
To identify the genes that have an elevated likelihood of contributing to ADHD pathology, we removed all genes duplicated with an incomplete promoter or coding sequence, or those aberrations found in any control subjects across all studies (Figure 1). Genes in rare CNVs identified in at least two patients were placed among the high-ranking candidates, because of their recurring nature. In addition, to narrow down the region of interest, we calculated the minimal region common to overlapping CNVs. The selection of the minimal chromosomal region is illustrated in Figure 1C. Together, this resulted in a high-ranking list of 432 genes (see Table S2 in the online supplement, high-priority gene list). The remaining 1,316 genes, observed in only a single patient, were considered low ranking (see Table S2, low-priority gene list).
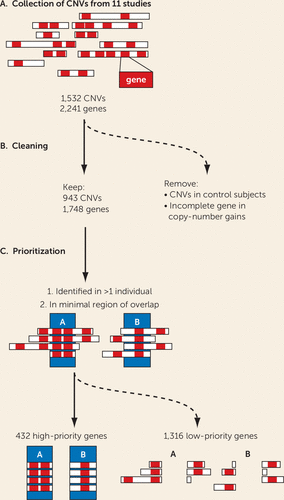
FIGURE 1. CNV processing scheme to establish high- and low-priority ADHD candidate genesa
a In panel A, copy number variant (CNV) coordinates were extracted from 11 studies reporting rare CNVs in attention deficit hyperactivity disorder (ADHD) cohorts (see Table S1 in the online supplement). In panel B, CNVs identified in control subjects and copy number gains with an incomplete promoter and coding sequences were excluded. In panel C, a high-priority gene list was generated by excluding genes found only in a single individual with ADHD and genes not present in the minimal overlapping region.
High-Priority ADHD Candidate Genes Show Increased Co-Expression in the Brain
It has been shown that proteins encoded by genes implicated in a genetically heterogeneous disorder tend to operate in common molecular pathways and processes (21, 32–35). To evaluate biological coherence of high-priority ADHD candidate genes in an unbiased way, we assessed their coexpression, a prerequisite for genes to jointly act in biological and developmental processes, during the development of the most relevant tissue, the brain. We used the BrainSpan data set to test for gene coexpression and found a significant enrichment (E) of coexpressed genes in the high-priority gene list (N=432; E=1.04, p=0.0044). The low-priority genes (N=1,316; E=1.01, p=0.28) did not show significant coexpression enrichment.
Cross-Species Phenotypes Link a Network of 66 High-Priority Candidate Genes to ADHD Core Symptoms
We used the Monarch Initiative cross-species genotype-phenotype database, which contains phenotypic information from 58 species, to 1) evaluate our gene prioritization, 2) retrieve independent evidence for the relevance of our high-priority candidate gene set for core ADHD features, and 3) identify functionally associated networks of high-priority genes with the ADHD core symptoms of hyperactivity, attention deficit, and impulsivity (for exact search terms, see Table S3 in the online supplement) (25). Eighteen of the 432 high-priority ADHD CNV candidate genes were associated with cross-species terms related to attention and hyperactivity: attention deficit hyperactivity disorder, hyperactivity, increased vertical activity, hyperactive, and abnormally increased process quality locomotory exploration behavior (Figure 2). While we could not directly test enrichment here, in a similar analysis we performed earlier (36), focused on Drosophila, we were able to show that orthologs of ADHD-associated genes have significantly more face-valid behaviors than randomly selected sets of genes, meaning that ADHD-associated human candidate genes occur more frequently among genes known to cause face-valid ADHD behaviors.
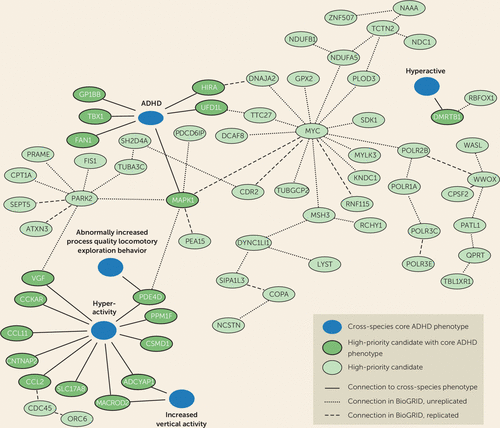
FIGURE 2. Cross-species phenotypes link an interaction network of 66 high-priority candidate genes to ADHD core symptomsa
a Of the 432 high-priority attention deficit hyperactivity disorder (ADHD) copy number variant (CNV) candidate genes, 18 were found in the Monarch Initiative database to have a core ADHD phenotype relating to hyperactivity, attention deficit, and impulsivity. These 18 genes were used as seeds to create a BioGRID (Biological General Repository for Interaction Datasets) interaction network, using cross-species information on direct protein-protein interactions, genetic interactions, and predicted interactions. We found a highly interconnected network with 48 secondary interactors from the high-priority gene list. Dashed lines show whether a connection was found once (unreplicated) or multiple times (replicated) in the BioGRID database.
Based on the 18 genes validated by the cross-species approaches, we mined the cross-species BioGRID database for interactors. This approach connected 48 additional genes of our high-priority catalog to the cross-species terms that are “guilty by association” (Figure 2). These genes have not yet been annotated with face-valid behaviors and should be prioritized for testing in such paradigms.
CNV-Derived High-Priority ADHD Candidate Genes Form Molecular Modules Implicating Specific Biological Processes in ADHD Pathology
In addition to the cross-species approach, we also used the DAPPLE algorithm to identify significantly connected proteins among the 432 high-priority genes. The DAPPLE analysis is based on the integration of experimentally validated protein-protein interaction databases to identify direct and indirect networks based on network and protein connectivity scores. The results of the algorithm depicted 17 modules of connected proteins, each comprising 2–15 ADHD high-priority candidates that directly interacted with each other (Figure 3). Taking both direct and indirect interactions into account, the hubs with significant connectivity contained 20 proteins (Figure 3; see also Table S4 in the online supplement). Of those, eight significant proteins were found in four direct protein-interaction modules (Figure 3, modules 1–4). Of those modules, 10 proteins—WWOX, PPM1F, PARK2, TUBA3C, MAPK1, MYC, SEPT5, POLR2B, POLR1A, and POLR3C—were found connected to cross-species core ADHD phenotypes (Figure 2).
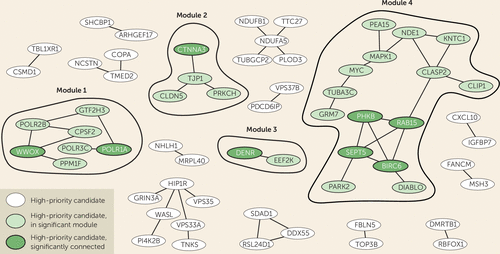
FIGURE 3. DAPPLE interaction network identifies four molecular modules with significantly connected proteinsa
a A Disease Association Protein-Protein Link Evaluator (DAPPLE) analysis was performed to identify highly related protein-protein interaction clusters based on direct and indirect connectivity. Four modules containing significantly connected proteins were identified.
The 26 Most Credible Genes Are Associated With Psychiatric Disorders in Common Variant GWASs
By integrating the information obtained from our different analytic approaches, we derived a “credible gene set.” This set of 26 genes among the original 432 high-priority CNV-derived ADHD candidate genes was consistently observed in all the different approaches we employed (Figure 4, Table 1). These 26 genes also showed significant enrichment of coexpressed genes (E=1.17, p=0.017), at a level higher than the entire high-priority list (E=1.04; see above).
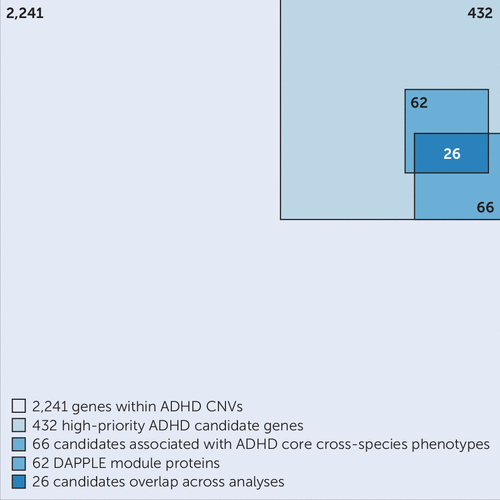
FIGURE 4. Visual representation of the gene selection processa
a A summary of the analysis process undertaken in the study, starting from 2,241 genes prioritized to 432 high-priority candidates (Figure 1), followed by a cross-species phenotype and BioGRID (Biological General Repository for Interaction Datasets) analysis of 66 genes (Figure 2) and a DAPPLE (Disease Association Protein-Protein Link Evaluator) analysis with 62 candidates (Figure 3). In total, 26 candidates overlap across analyses (Table 1). ADHD=attention deficit hyperactivity disorder; CNV=copy number variant.
We obtained additional, complementary evidence for the relevance of these 26 genes for ADHD with yet another source of data, namely, common genetic variants as retrieved from the ADHD GWAS summary statistics. This specific stepwise analysis approach allowed us to prioritize only the best candidate genes for a role in ADHD pathophysiology supported by evidence from multiple sources. We performed gene-based association analyses for all 26 genes to evaluate whether they are also implicated in ADHD risk through common genetic variants, using the largest meta-analytic GWAS data currently available for ADHD (N=55,374, PGC-iPSYCH ADHD Working Group). Two genes were significantly associated with ADHD after correction for multiple testing: POLR3C (p=0.000020) and RBFOX1 (p=0.00018) (see Table S5 and Figure S1 in the online supplement). Interestingly, POLR3C was among the top 0.43% of the most strongly associated genes (ranking at 80 of 18,411). When investigating the specificity of the findings for ADHD, we found that five of the 26 genes individually also showed significant gene-based association with a combined measure across eight psychiatric disorders (p<0.002 for RBFOX1, MAPK1, POLR3C, CSMD1, PPM1F; see Table S6 in the online supplement). Gene expression heat maps were constructed to show the coexpression pattern of the 26 genes in all tissues and in brain from developing to adult stages. As shown in Figure S2 in the online supplement, over half of the genes show a broad expression pattern across different tissue types, while the others are more restricted in brain (e.g., RBFOX1 and CSMD1). The developmental analysis showed that about half of the 26 genes are expressed from early prenatal development into adulthood, while most others show expression patterns more restricted to prenatal development.
Discussion
Here, we present an integrated analysis of ADHD-associated CNVs identified in the 11 studies published to date that have detected rare CNVs in ADHD case subjects. The limited power of the individual studies has been a crucial bottleneck in the definition of high-priority ADHD candidate genes for further studies. From the 1,532 CNVs described in the 11 studies, using strict criteria, we extracted 2,241 mRNA-coding genes; this number is likely to contain many false positives because of the individually low occurrence of the rare CNVs (19), but the number is too large to allow each CNV to be studied in (animal) models for validation and mechanistic insights. In this study, we aimed to prioritize genes linked to ADHD among the 2,241 on the basis of the robustness of findings across different bioinformatics approaches, used in an integrative manner and including both human and animal model–derived data. For this, we selected only those genes that were recurrently affected by CNVs in the case subjects and focused on the minimal overlapping region of different CNVs in a region. Furthermore, we excluded all genes that were affected by a CNV in healthy control subjects of other studies, as well as those that were only partially duplicated (lacking a full coding sequence and promoter). These stringent criteria substantially reduced the number of candidate genes from the CNVs, to one-fifth of the original number. We showed that the selected 432 high-priority genes were significantly more coexpressed in the developing brain in comparison to random gene groups; this is evidence that our selection enriches for biologically coherent genes that are expressed at the same time in the same tissue, a prerequisite for them to be involved in the same biological processes and cause similar phenotypes when disturbed.
The contribution of rare genetic variation, including rare CNVs, to ADHD pathophysiology has received relatively limited attention thus far. Given the high prevalence of the disorder, the focus of genetic studies has been largely on common genetic variants, following the “common disease–common variant” hypothesis (37, 38). However, in most common diseases, rare variants have also been shown to play a role, which has prompted researchers to start studying them for ADHD as well. Most studies published thus far have been limited in both sample size and in their focus on only one form of rare variants—CNVs—requiring integration as performed in the present study to extract meaning. Recent work in larger samples and/or using alternative methods for rare variant detection (e.g., exome sequencing) confirmed a role of such rare variation in ADHD. Most notably, the large study in the iPSYCH sample (39) confirmed a significantly higher burden of rare protein-truncating variants in evolutionarily constrained genes in ADHD case subjects than in control subjects. Also, a recent CNV study in over 400 patients identified rare CNVs in 9.4% of individuals with ADHD (40), and two studies of parent-offspring trios pinpointed several de novo CNVs occurring in offspring with ADHD (41, 42). Whether these rare variants are the sole (genetic) cause of ADHD in the individual carrying them remains to be clarified. So far, evidence supporting high penetrance is been limited (43, 44). Irrespective of this, the fact that rare variants can often be more easily linked to gene dysfunction than common genetic variants makes them an invaluable source of information toward understanding the biology underlying ADHD.
Studies in model organisms can provide a wealth of phenotypic information, because of the high level of functional conservation across species. For ADHD, several model organisms have been shown to provide valid phenotypes. These include monkey, rat, mouse, zebrafish, and fruit fly, where relevant phenotypes can be observed on genetic manipulation or drug administration (6). To identify biological processes underlying the selection of genes, we therefore took a novel approach that has not yet been applied in the field of neuropsychiatric disorders. We mined the Monarch Initiative database, which integrates genotype-phenotype relations across species. Using this approach, we found that 18 of the 432 high-priority genes, when manipulated in animal models, cause phenotypes that are face valid to the ADHD core phenotypes of attention deficit, hyperactivity, and impulsivity. The beauty of animal models is that paradigms can be tested robustly and under controlled conditions, with large numbers of genetically identical animals and under environmentally identical conditions. This is impossible to achieve in humans. Although animal models are not a copy of humans, there is ample scientific evidence that fundamental behaviors such as learning, memory, and circadian and activity/sleep rhythms rely on highly conserved molecular mechanisms and players (45, 46). Importantly, ADHD has consistently been shown to represent the extreme of traits of activity and attention in the general population; this has been established on the phenotypic, genetic, and neuroimaging levels (7, 47). This fact makes ADHD one of the psychiatric disorders that can be modeled particularly well in animal models. Our approach integrating across model systems is therefore a particular strength of this study, with the consistency of our findings across the different approaches illustrating its value.
It is highly likely that the proteins we identified using the Monarch Initiative database form functional biological connections with proteins for which detailed functional characterization, in particular information on ADHD-related phenotypes, is still lacking. We therefore retrieved direct interactors of the 18 proteins and identified an interconnected network of 66 proteins linked directly or indirectly to disease-relevant phenotypes (Figure 2). We also tested whether our high-priority gene list itself formed protein-interaction modules with significantly interconnected proteins, which could provide us with information on biological processes. Indeed, we identified four modules comprising significantly connected proteins from our selection. Of these, the major modules, module 1 and module 4, connected to and were supported by ADHD-related phenotypes across species, showing the added value of cross-species analysis (Figure 2).
Module 1 contained two significantly linked proteins: WW-domain containing oxidoreductase (WWOX) and DNA-directed RNA polymerase I subunit RPA1 (POLR1A). WWOX is involved in autosomal recessive cerebellar ataxia-epilepsy-intellectual disability syndrome (48), but no direct connections with ADHD are known. However, WWOX interacts with protein phosphatase 1F (PPM1F), a Ser/Thr protein phosphatase that modulates RhoA and Ca2+/calmodulin–dependent protein kinase II pathways (49, 50). Other members of this phosphatase protein family are involved in mediating dopaminergic signaling via G-protein-coupled receptors (51). In addition, misexpression of PPM1F in the substantia nigra in patients with Parkinson’s disease may implicate PPM1F more directly in dopaminergic biology (52). Dopamine signaling pathways have repeatedly been found to be altered in ADHD patients, and they form the basis for the most widely used pharmacological treatment approach (53, 54). As we can directly connect PPM1F to the cross-species term hyperactivity and indirectly link WWOX, POLR1A, POLR2B, and POLR3C to ADHD core phenotypes, we can extend the network with genes potentially regulating dopaminergic signaling (Figure 2). The RNA polymerase II subunits (POLR1A, POLR2B, and POLR3C) are involved in the regulation and fine-tuning of transcription.
Module 2 clusters proteins that are required for blood-brain barrier formation, which function in cell-cell junctions and communication. This module contains one significantly connected protein: catenin alpha-3 (CTNNA3). This adherence junction protein, also known to be associated with autism spectrum disorder (ASD), likely modulates cerebral and ependymal regions through GABAA receptor activation (55). Tight junction protein ZO-1 (TJP1) forms the connection to the other three proteins in this module. This gene is affected by CNVs in 28 ADHD case subjects, being the most frequently occurring gene affected by copy number alterations in our survey of the ADHD CNV studies (see Table S1 in the online supplement). TJP1, together with claudin-5 (CLDN5, affected in 12 ADHD case subjects) represents an important constituent of the blood-brain barrier (56, 57). Protein kinase C eta type (PRKCH) regulates TJP1 (58). Genes regulating neuronal cell adhesion are also significantly associated with ASD, schizophrenia, and bipolar disorder, raising the hypothesis that this mechanism plays a role across different neuropsychiatric disorders (59, 60). Given the high number of ADHD case subjects with a CNV in this module, we postulate that cell-cell junctions play an important role in ADHD.
Module 3 contains two proteins that directly interact with each other: the significantly interconnected density-regulated protein (DENR) and the eukaryotic elongation factor 2 kinase (EEF2K), both involved in regulation and initiation of translation (61). Regulation and initiation of translation have been linked to neuropsychiatric disorders, including ADHD, through the regulation of brain-derived neurotrophic factor (62–64). Based on the repeated association and the described functional work, we suggest transcriptional regulation as one of the mechanisms that modulate the risk for ADHD.
Module 4 contains four significantly connected proteins: BIRC6, RAB15, SEPT5, and PHKB. Baculoviral IAP repeat-containing protein 6 (BIRC6 or Bruce) is an inhibitor of apoptosis involved in prostate cancer progression, but it also acts in neuronal protection against apoptosis (65, 66). Ras-related protein Rab-15 (RAB15) is a direct connector of BIRC6, and it plays a role in regulating synaptic vesicle membrane flow in nerve terminals (67, 68). Septin-5 (SEPT5) is involved in the binding of SNARE complexes, inhibiting synaptic vesicle exocytosis (69). Recent studies have shown that manipulation of this gene in mice leads to altered social interaction and altered affective behaviors (70, 71). Phosphorylase b kinase regulatory subunit beta (PHKB) is involved in glycogen metabolism and has been linked to neuronal plasticity (72). Other proteins in the hub link to the cross-species phenotype term hyperactivity and attention deficit hyperactivity disorder: MAPK1 is directly connected, and SEPT5, PARK2, MYC, and TUBA3C are indirectly connected. They are thus prime candidates for further evaluation in functional assays.
While this study started from rare CNVs, we also found corroborating evidence for several of the genes implicated in ADHD in studies of common genetic variants. Among the 26 CNV-affected genes most consistently observed across the different bioinformatics approaches applied in this study, we also found RBFOX1 and POLR3C to be associated with the disorder in the largest SNP-based GWAS meta-analysis to date. In a recent study by Lee et al. (73), RBFOX1 was discovered to be the second most pleiotropic locus of the genome-wide meta-analysis among eight psychiatric disorders. RBFOX1 encodes a splice regulator regulating several genes involved in neuronal development and mainly expressed in the brain (73–76). Animal models have shown that RBFOX1 is involved in mouse corticogenesis and aggressive behaviors (73, 75, 77, 78). POLR3C is included in a well-known small CNV located at 1q21.1, which contributes to a broad spectrum of phenotypes in addition to ADHD, including morphological features and ASD (79, 80). The 26 genes listed in Table 1 may be viewed as having, individually, the highest credibility as ADHD candidate genes. We therefore recommend that these genes be prioritized in future studies searching for rare (single-nucleotide) variants in ADHD and for functional characterization of gene-disease pathways. Table 1 also shows that among these 26 genes, common biological themes are present, such as transcription (already highlighted by the module analysis), mitochondria biology, mRNA metabolism, and cytoskeleton.
It is likely that the gene modules we identified for ADHD are also relevant for other neurodevelopmental disorders. Several of the individual studies that formed the basis for our work already reported overlap of identified CNVs and copy-variant genes with those found associated with ASD, intellectual disability, and schizophrenia (e.g., 8, 18). Our analysis of the 26 top genes in cross-disorder GWAS data from eight different psychiatric disorders also suggested pleiotropy. Whether the gene-gene interactions in the specific modules may be more relevant for ADHD than for the other disorders will need to be clarified in future work.
Analysis of the expression patterns of the 26 most credible genes revealed a varied expression, with several genes being restricted to the brain and others showing a broad expression pattern across different tissue types. The developmental analysis showed the most overlap between genes during prenatal development. We were surprised to see that TUBA3C and DMRTB1 appeared to show expression restricted to the testis in the GTEx data and also did not show brain expression in the BrainSpan data. However, this could be a technical/analytical artifact, as the TUBA3C gene is known to be mutated in Kabuki syndrome, a genetic syndrome including intellectual disability, and DMRTB1 was earlier identified as an interactor of, for example, RBFOX1 in a screen for protein-protein interactions relevant in inherited neurodegenerative disorders (81). We did not observe restriction of gene expression to specific brain regions. This is consistent with results from recent brain imaging studies, where structural alterations in patients with ADHD were found to be a rather global phenomenon affecting multiple brain measures, with the highest effect sizes found for global measures, such as intracranial volume (a proxy for total brain volume) (47, 82).
Our study should be viewed in the light of some strengths and limitations. We show that filtering based on recurrent CNVs restricted to ADHD case subjects in conjunction with complementary bioinformatics methods bear great potential to prioritize ADHD candidate genes. For the first time, cross-species phenotypes were used to identify candidate genes linked to ADHD core phenotypes, pointing to high-priority candidate genes. Several of the identified genes form significantly connected protein networks characterized by shared functions. In the cross-species analyses, we found that 56% of the genes (684/1,213; see Table S3 in the online supplement) were annotated with the object label “hyperactivity”; this overrepresentation may result from the fact that activity is more easily assessed in animal models than are cognitive phenotypes, such as attention and impulsivity. The selection of genes analyzed in this study is extensive but by no means exhaustive. First, it is important to note that by only analyzing recurrent CNVs and focusing on the minimal regions of overlap among CNVs in the same region, genes with relevance for ADHD may be overlooked; on the other hand, the stringent evidence-based filtering holds high potential to uncover the most ADHD-relevant biological pathways. In addition, neither the surveyed CNV studies nor our study consider effects of CNVs on the surrounding genetic landscape. A duplicated CNV translocation, for example, can have an impact on the expression of genes in the chromosomal region at the site of insertion, which alone or together with the duplicated genes can contribute to ADHD phenotypes (83). An additional limitation was the fact that the criteria used for selection of CNVs were not identical across the 11 source studies. Furthermore, the primary studies included here evaluated only autosomal CNVs; given the known sex differences in the prevalence of ADHD, it will be interesting to extend such work to the sex chromosomal CNVs. Lastly, we used a concatenation approach across brain regions and developmental time points based on the BrainSpan data set in our bioinformatic analysis, in keeping with the broad involvement of cortical and subcortical brain regions in ADHD (47, 82). Knowing that not all brain regions and developmental stages are well represented in the BrainSpan data, we may have missed specificity in the coexpression modules. While we chose for maximal power through concatenation, ideally, one would also want to consider spatial and temporal gene expression patterns independently. With the more comprehensive data sets now becoming available, opportunities for such analyses will improve for future studies (84, 85).
Conclusions
Our study shows that stringent filtering in CNV studies in combination with a complementary battery of bioinformatics approaches can identify ADHD candidate genes at increased levels of credibility. We suggest that testing genes operating in the identified modules and the 26 genes consistently found in all approaches employed here in animal models such as rat, mouse, zebrafish, and fruit fly (6) for their ability to modulate behavior will provide further insights into the mechanistic pathways and biology of ADHD and potentially other neurodevelopmental disorders. It is clear that such a deeper understanding of ADHD is essential for improving its clinical management. This includes the development of novel treatment approaches, the development of biomarkers and diagnostic tools for the disorder, and potentially informing improvements in the nosology of psychiatric disorders toward more biologically defined disease definitions.
1 : The worldwide prevalence of ADHD: a systematic review and metaregression analysis. Am J Psychiatry 2007; 164:942–948Link, Google Scholar
2 : The genetics of attention deficit/hyperactivity disorder in adults, a review. Mol Psychiatry 2012; 17:960–987Crossref, Medline, Google Scholar
3 : Attention-deficit/hyperactivity disorder. Nat Rev Dis Primers 2015; 1:15020Crossref, Medline, Google Scholar
4 : Candidate gene studies of ADHD: a meta-analytic review. Hum Genet 2009; 126:51–90Crossref, Medline, Google Scholar
5 : Genome-wide association studies in ADHD. Hum Genet 2009; 126:13–50Crossref, Medline, Google Scholar
6 : Brain imaging genetics in ADHD and beyond: mapping pathways from gene to disorder at different levels of complexity. Neurosci Biobehav Rev 2017; 80:115–155Crossref, Medline, Google Scholar
7 : Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet 2019; 51:63–75Crossref, Medline, Google Scholar
8 : Genome-wide analysis of copy number variants in attention deficit hyperactivity disorder: the role of rare variants and duplications at 15q13.3. Am J Psychiatry 2012; 169:195–204Link, Google Scholar
9 : Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet 2010; 376:1401–1408Crossref, Medline, Google Scholar
10 : Polygenic transmission and complex neuro developmental network for attention deficit hyperactivity disorder: genome-wide association study of both common and rare variants. Am J Med Genet Part B Neuropsychiatr Genet 2013; 162:419–430Crossref, Google Scholar
11 : Investigating the contribution of common genetic variants to the risk and pathogenesis of ADHD. Am J Psychiatry 2012; 169:186–194Link, Google Scholar
12 : Genome-wide copy number variation analysis in adult attention-deficit and hyperactivity disorder. J Psychiatr Res 2014; 49:60–67Crossref, Medline, Google Scholar
13 : Biological overlap of attention-deficit/hyperactivity disorder and autism spectrum disorder: evidence from copy number variants. J Am Acad Child Adolesc Psychiatry 2014; 53:761–770.e26Crossref, Medline, Google Scholar
14 : Rare copy number variation discovery and cross-disorder comparisons identify risk genes for ADHD. Sci Transl Med 2011; 3:95ra75Crossref, Medline, Google Scholar
15 : Genome-wide copy number variation analysis in attention-deficit/hyperactivity disorder: association with neuropeptide Y gene dosage in an extended pedigree. Mol Psychiatry 2011; 16:491–503Crossref, Medline, Google Scholar
16 : Genome-wide analysis of rare copy number variations reveals PARK2 as a candidate gene for attention-deficit/hyperactivity disorder. Mol Psychiatry 2014; 19:115–121Crossref, Medline, Google Scholar
17 : Genome-wide copy number variation study associates metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nat Genet 2012; 44:78–84Crossref, Google Scholar
18 : Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry 2010; 15:637–646Crossref, Medline, Google Scholar
19 : Interpreting the role of de novo protein-coding mutations in neuropsychiatric disease. Nat Genet 2013; 45:234–238Crossref, Medline, Google Scholar
20 : The discovery of integrated gene networks for autism and related disorders. Genome Res 2015; 25:142–154Crossref, Medline, Google Scholar
21 : Psychiatric gene discoveries shape evidence on ADHD’s biology. Mol Biology 2016; 21:1202–1207Google Scholar
22 : Phenotypic overlap in the contribution of individual genes to CNV pathogenicity revealed by cross-species computational analysis of single-gene mutations in humans, mice, and zebrafish. Dis Model Mech 2013; 6:358–372Crossref, Medline, Google Scholar
23 : Construction and accessibility of a cross-species phenotype ontology along with gene annotations for biomedical research. F1000Research 2013; 2:30Crossref, Medline, Google Scholar
24 : An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 2012; 489:391–399Crossref, Medline, Google Scholar
25 : The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res 2017; 45(D1):D712–D722Crossref, Medline, Google Scholar
26 : Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2007; 2:2366–2382Crossref, Medline, Google Scholar
27 : Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet 2011; 7:e1001273Crossref, Medline, Google Scholar
28 : GenePattern 2.0. Nat Genet 2006; 38:500–501Crossref, Medline, Google Scholar
29 : Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell 2019; 179:1469–1482Crossref, Medline, Google Scholar
30 : MAGMA: generalized gene-set analysis of GWAS data. PLOS Comput Biol 2015; 11:e1004219Crossref, Medline, Google Scholar
31 : Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017; 8:1826Crossref, Medline, Google Scholar
32 : Psychiatric genome-wide association study analyses implicate neuronal, immune, and histone pathways. Nat Neurosci 2015; 18:199–209Crossref, Medline, Google Scholar
33 : Pathway analysis in attention deficit hyperactivity disorder: an ensemble approach. Am J Med Genet Part B Neuropsychiatr Genet 2016; 171:815–826Crossref, Medline, Google Scholar
34 : Characterizing autism spectrum disorders by key biochemical pathways. Front Neurosci 2015; 9:313Crossref, Medline, Google Scholar
35 : Systematic phenomics analysis deconvolutes genes mutated in intellectual disability into biologically coherent modules. Am J Hum Genet 2016; 98:149–164Crossref, Medline, Google Scholar
36 : ADHD-associated dopamine transporter, latrophilin and neurofibromin share a dopamine-related locomotor signature in Drosophila. Mol Psychiatry 2016; 21:565–573Crossref, Medline, Google Scholar
37 : On the allelic spectrum of human disease. Trends Genet 2001; 17:502–510Crossref, Medline, Google Scholar
38 : Common vs rare allele hypotheses for complex diseases. Curr Opin Genet Dev 2009; 19:212–219Crossref, Medline, Google Scholar
39 : Autism spectrum disorder and attention deficit hyperactivity disorder have a similar burden of rare protein-truncating variants. Nat Neurosci 2019; 22:1961–1965Crossref, Medline, Google Scholar
40 : A large data resource of genomic copy number variation across neurodevelopmental disorders. NPJ Genom Med 2019; 4:26Crossref, Medline, Google Scholar
41 : A brief report: de novo copy number variants in children with attention deficit hyperactivity disorder. Transl Psychiatry 2020; 10:135Crossref, Medline, Google Scholar
42 : Sequencing of sporadic attention-deficit hyperactivity disorder (ADHD) identifies novel and potentially pathogenic de novo variants and excludes overlap with genes associated with autism spectrum disorder. Am J Med Genet B Neuropsychiatr Genet 2017; 174:381–389Crossref, Medline, Google Scholar
43 : Quantifying the impact of rare and ultra-rare coding variation across the phenotypic spectrum. Am J Hum Genet 2018; 102:1204–1211Crossref, Medline, Google Scholar
44 : Identification of ADHD risk genes in extended pedigrees by combining linkage analysis and whole-exome sequencing. Mol Psychiatry (Epub ahead of print, August 16, 2018)Google Scholar
45 : Time for bed: genetic mechanisms mediating the circadian regulation of sleep. Trends Genet 2018; 34:379–388Crossref, Medline, Google Scholar
46 : 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nat Rev Neurosci 2010; 11:514–522Crossref, Medline, Google Scholar
47 : Brain imaging of the cortex in ADHD: a coordinated analysis of large-scale clinical and population-based samples. Am J Psychiatry 2019; 176:531–542Link, Google Scholar
48 : The tumour suppressor gene WWOX is mutated in autosomal recessive cerebellar ataxia with epilepsy and mental retardation. Brain 2014; 137:411–419Crossref, Medline, Google Scholar
49 : Regulation of the multifunctional Ca2+/calmodulin-dependent protein kinase II by the PP2C phosphatase PPM1F in fibroblasts. J Biol Chem 2004; 279:24889–24898Crossref, Medline, Google Scholar
50 : Functional interactions between phosphatase POPX2 and mDia modulate RhoA pathways. J Cell Sci 2008; 121:514–521Crossref, Medline, Google Scholar
51 : Dopamine D2 receptor relies upon PPM/PP2C protein phosphatases to dephosphorylate huntingtin protein. J Biol Chem 2014; 289:11715–11724Crossref, Medline, Google Scholar
52 : Gene expression profiling of substantia nigra dopamine neurons: further insights into Parkinson’s disease pathology. Brain 2009; 132:1795–1809Crossref, Medline, Google Scholar
53 : Dopamine genes and ADHD. Neurosci Biobehav Rev 2005; 24:21–25Crossref, Google Scholar
54 : Imaging the effects of methylphenidate on brain dopamine: new model on its therapeutic actions for attention-deficit/hyperactivity disorder. Biol Psychiatry 2005; 57:1410–1415Crossref, Medline, Google Scholar
55 : αT-catenin in restricted brain cell types and its potential connection to autism. J Mol Psychiatry 2016; 4:2Crossref, Medline, Google Scholar
56 : Zebrafish as a model for developmental neurotoxicity assessment: the application of the zebrafish in defining the effects of arsenic, methylmercury, or lead on early neurodevelopment. Toxics 2014; 2:464–495 (https://doi.org/10.3390/toxics2030464)Crossref, Google Scholar
57 : Tight junction modulation of the blood brain barrier: CNS delivery of small molecules. Tissue Barriers 2016; 4:e1138017Crossref, Medline, Google Scholar
58 : PKC eta regulates occludin phosphorylation and epithelial tight junction integrity. Proc Natl Acad Sci USA 2009; 106:61–66Crossref, Medline, Google Scholar
59 : Association of the neuronal cell adhesion molecule (NRCAM) gene variants with autism. Int J Neuropsychopharmacol 2009; 12:1–10Crossref, Medline, Google Scholar
60 : Molecular pathways involved in neuronal cell adhesion and membrane scaffolding contribute to schizophrenia and bipolar disorder susceptibility. Mol Psychiatry 2011; 16:286–292Crossref, Medline, Google Scholar
61 : cdc2-cyclin B regulates eEF2 kinase activity in a cell cycle‐ and amino acid‐dependent manner. EMBO J 2008; 27:1005–1016Crossref, Medline, Google Scholar
62 : The neurology of mTOR. Neuron 2014; 84:275–291Crossref, Medline, Google Scholar
63 : NMDA receptor blockade at rest triggers rapid behavioural antidepressant responses. Nature 2011; 475:91–95Crossref, Medline, Google Scholar
64 : Rare DNA variants in the brain-derived neurotrophic factor gene increase risk for attention-deficit hyperactivity disorder: a next-generation sequencing study. Mol Psychiatry 2017; 22:580–584Crossref, Medline, Google Scholar
65 : BIRC6 protein, an inhibitor of apoptosis: role in survival of human prostate cancer cells. PLoS One 2013; 8:e55837Crossref, Medline, Google Scholar
66 : Bruce/apollon promotes hippocampal neuron survival and is downregulated by kainic acid. Biochem Biophys Res Commun 2005; 338:729–735Crossref, Medline, Google Scholar
67 : rab15, a novel low molecular weight GTP-binding protein specifically expressed in rat brain. J Biol Chem 1992; 267:5768–5775Medline, Google Scholar
68 : Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 2009; 10:513–525Crossref, Medline, Google Scholar
69 : The septin Sept5/CDCrel-1 competes with alpha-SNAP for binding to the SNARE complex. Biochem J 2005; 385:347–353Crossref, Medline, Google Scholar
70 : Sept5 deficiency exerts pleiotropic influence on affective behaviors and cognitive functions in mice. Hum Mol Genet 2009; 18:1652–1660Crossref, Medline, Google Scholar
71 : Alterations of social interaction through genetic and environmental manipulation of the 22q11.2 gene Sept5 in the mouse brain. Hum Mol Genet 2012; 21:3489–3499Crossref, Medline, Google Scholar
72 : Learning-induced gene expression in the hippocampus reveals a role of neuron-astrocyte metabolic coupling in long term memory. PLoS One 2015; 10:e0141568Crossref, Medline, Google Scholar
73 : Genome wide meta-analysis identifies genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell 2019; 179:1469–1482.e11Crossref, Medline, Google Scholar
74 : The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet 2011; 43:706–711Crossref, Medline, Google Scholar
75 : Role of the cytoplasmic isoform of RBFOX1/A2BP1 in establishing the architecture of the developing cerebral cortex. Mol Autism 2015; 6:56Crossref, Medline, Google Scholar
76 : Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 2018; 359:693–697Crossref, Medline, Google Scholar
77 : Essential role of the nuclear isoform of RBFOX1, a candidate gene for autism spectrum disorders, in the brain development. Sci Rep 2016; 6:30805Crossref, Medline, Google Scholar
78 : RBFOX1, encoding a splicing regulator, is a candidate gene for aggressive behavior. Eur Neuropsychopharmacol 2020; 30:44–55Crossref, Medline, Google Scholar
79 : Expanding the phenotype of reciprocal 1q21.1 deletions and duplications: a case series. Ital J Pediatr 2017; 43:61Crossref, Medline, Google Scholar
80 : Variants in the 1q21 risk region are associated with a visual endophenotype of autism and schizophrenia. Genes Brain Behav 2014; 13:144–151Crossref, Medline, Google Scholar
81 : A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell 2006; 125:801–814Crossref, Medline, Google Scholar
82 : Subcortical brain volume differences in participants with attention deficit hyperactivity disorder in children and adults: a cross-sectional mega-analysis. Lancet Psychiatry 2017; 4:310–319Crossref, Medline, Google Scholar
83 : Non-coding genetic variants in human disease. Hum Mol Genet 2015; 24(R1):R102–R110Crossref, Medline, Google Scholar
84 : Brain transcriptome databases: a user’s guide. J Neurosci 2018; 38:2399–2412Crossref, Medline, Google Scholar
85 : Whole-genome and RNA sequencing reveal variation and transcriptomic coordination in the developing human prefrontal cortex. Cell Rep 2020; 31:107489PubMedCrossref, Google Scholar



