Lower Gene Expression for KCNS3 Potassium Channel Subunit in Parvalbumin-Containing Neurons in the Prefrontal Cortex in Schizophrenia
Abstract
Objective
In schizophrenia, alterations in markers of cortical GABA neurotransmission are prominent in parvalbumin-containing neurons. Parvalbumin neurons selectively express KCNS3, the gene encoding the Kv9.3 potassium channel α-subunit. Kv9.3 subunits are present in voltage-gated potassium channels that contribute to the precise detection of coincident excitatory synaptic inputs to parvalbumin neurons. This distinctive feature of parvalbumin neurons appears important for the synchronization of cortical neural networks in γ-oscillations. Because impaired prefrontal cortical γ-oscillations are thought to underlie the cognitive impairments in schizophrenia, the authors investigated whether KCNS3 mRNA levels are altered in the prefrontal cortex of schizophrenia subjects.
Method
KCNS3 mRNA expression was evaluated by in situ hybridization in 22 matched pairs of schizophrenia and comparison subjects and by microarray analyses of pooled samples of individually dissected neurons that were labeled with Vicia villosa agglutinin (VVA), a parvalbumin neuron-selective marker, in a separate cohort of 14 pairs. Effects of chronic antipsychotic treatments on KCNS3 expression were tested in the prefrontal cortex of antipsychotic-exposed monkeys.
Results
By in situ hybridization, KCNS3 mRNA levels were 23% lower in schizophrenia subjects. At the cellular level, both KCNS3 mRNA-expressing neuron density and KCNS3 mRNA level per neuron were significantly lower. By microarray, KCNS3 mRNA levels were lower by 40% in VVA-labeled neurons from schizophrenia subjects. KCNS3 mRNA levels were not altered in antipsychotic-exposed monkeys.
Conclusions
These findings reveal lower KCNS3 expression in prefrontal cortical parvalbumin neurons in schizophrenia, providing a molecular basis for compromised detection of coincident synaptic inputs to parvalbumin neurons that could contribute to altered γ-oscillations and impaired cognition in schizophrenia.
Cognitive deficits, which represent a treatment-resistant, disabling symptom domain in schizophrenia, are attributable, at least in part, to alterations in cortical interneurons that utilize GABA as an inhibitory neurotransmitter (1–4). These alterations appear to be prominent in the subset of GABA neurons that express parvalbumin. For example, in the prefrontal cortex of subjects with schizophrenia, parvalbumin neurons exhibit lower mRNA and protein levels of GAD67 (the 67-kDa isoform of glutamic acid decarboxylase, an enzyme for GABA synthesis) (5, 6), reduced immunoreactivity of GABA transporter 1 (a presynaptic GABA transporter) (7), and lower parvalbumin mRNA expression (5, 8–10). Because parvalbumin neurons are essential for the generation of γ-oscillations that appear to be a neural substrate for cognitive functions (11, 12), the alterations in parvalbumin neurons are thought to contribute to γ-oscillation disturbances and cognitive deficits in schizophrenia (3, 13).
We recently demonstrated (14) the selective expression of KCNS3, the gene encoding Kv9.3 voltage-gated potassium channel modulatory α-subunit, in parvalbumin neurons in the human prefrontal cortex. In heterologous expression systems, Kv9.3 subunits do not assemble into homomeric channels but form functional heteromeric channels with delayed rectifier Kv2.1 α-subunits (15–17), which are expressed by the majority of cortical neurons, including parvalbumin neurons (18). Compared with homomeric Kv2.1 channels, heteromeric Kv2.1/Kv9.3 channels have faster activation, slower deactivation and inactivation, and steady-state activation and inactivation curves that are shifted toward more negative values by ∼20 mV (15, 16). Therefore, Kv2.1/Kv9.3 channels appear to be more effectively activated by subthreshold membrane depolarizations such as those generated by excitatory synaptic inputs.
Parvalbumin neurons have a faster decay of excitatory postsynaptic potentials than other types of cortical neurons (19–21). Because fast potentials summate within a short time window (22), parvalbumin neurons fire action potentials only if they receive highly coincident excitatory inputs. Activation of voltage-gated potassium channels during excitatory postsynaptic potentials is one mechanism for the fast potentials in parvalbumin neurons (22). Therefore, the selective expression of KCNS3-encoded Kv9.3 subunits in parvalbumin neurons appears to contribute to the ability of parvalbumin neurons to detect coincident excitatory synaptic inputs that reflect synchronized cortical activities, and thus might contribute to their critical role in generating γ-oscillations.
Given the evidence for impaired γ-oscillations in schizophrenia, we sought to determine whether KCNS3 expression is altered in the prefrontal cortex of subjects with schizophrenia. Two complementary approaches in separate cohorts of subjects were used. In one cohort, KCNS3 mRNA was visualized by in situ hybridization and quantified at both the tissue and cellular levels. In the second cohort, parvalbumin neurons were identified by the presence of perineuronal nets and captured by laser microdissection; KCNS3 and KCNB1 (which encodes Kv2.1) mRNAs were measured by microarray.
Method
Human Subjects
Brain specimens were obtained during autopsies conducted at the Allegheny County Medical Examiner’s Office in Pittsburgh after consent was obtained from the next of kin. An independent committee of experienced research clinicians made consensus DSM-IV diagnoses for each subject using structured interviews with family members and medical records, as described previously (10); the same approach was used to confirm the absence of psychiatric diagnoses in comparison subjects. Two independent cohorts of subjects with schizophrenia and matched comparison subjects were used (Table 1). To control for biological and experimental variance, each schizophrenia subject was individually matched for sex and, as closely as possible, for age with one comparison subject (see Tables S1 and S2 in the data supplement that accompanies the online edition of this article). The first cohort (Table 1, Table S1), used for in situ hybridization studies, was composed of 22 subject pairs that did not differ significantly in age, postmortem interval, RNA integrity number (RIN; determined by the Agilent Bioanalyzer), brain pH, or tissue storage time. The second cohort, used for laser microdissection microarray studies (Table 1, Table S2), was composed of a separate set of 14 subject pairs that did not differ significantly in age, postmortem interval, RIN, or tissue storage time. However, comparison subjects had a significantly higher mean brain pH than schizophrenia subjects (6.7 [SD=0.16] compared with 6.4 [SD=0.20]; t=4.4, p<0.001).
| In Situ Hybridization | Microarray | |||||||
|---|---|---|---|---|---|---|---|---|
| Characteristic | Comparison Subjects (N=22) | Schizophrenia Subjects (N=22) | Comparison Subjects (N=14) | Schizophrenia Subjects (N=14) | ||||
| N | % | N | % | N | % | N | % | |
| Male | 17 | 77 | 17 | 77 | 10 | 71 | 10 | 71 |
| Race | ||||||||
| White | 18 | 82 | 14 | 64 | 12 | 86 | 10 | 71 |
| Black | 4 | 18 | 8 | 36 | 2 | 14 | 4 | 29 |
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Age (years) | 48.0 | 15.3 | 47.9 | 14.5 | 46.7 | 11.3 | 44.3 | 11.2 |
| Postmortem interval (hours) | 18.4 | 5.4 | 18.4 | 9.0 | 17.1 | 6.6 | 17.7 | 8.4 |
| Brain pH | 6.8 | 0.1 | 6.8 | 0.1 | 6.7 | 0.2 | 6.4 | 0.2 |
| RNA integrity number | 8.6 | 0.4 | 8.4 | 0.7 | 7.8 | 0.5 | 7.8 | 0.5 |
| Storage time (months at –80°C) | 161.2 | 32.8 | 171.9 | 23.7 | 91.7 | 22.4 | 85.2 | 9.6 |
All procedures were approved by the University of Pittsburgh’s Committee for the Oversight of Research Involving the Dead and Institutional Review Board for Biomedical Research, as well as by the Ethics Committee of Kanazawa University Graduate School of Medical Science.
In Situ Hybridization
The right hemisphere of each brain was blocked coronally, immediately frozen, and stored at −80°C (23). Cryostat sections of prefrontal cortex area 9 were cut serially from the rostral-caudal level corresponding to the middle portion of the superior frontal gyrus, as described previously (10). Some sections were collected into tubes containing TRIzol reagent (Invitrogen, Grand Island, N.Y.) for RNA isolation and subsequent RIN determination. For Nissl staining or in situ hybridization, 20-μm sections were mounted on Superfrost Plus glass slides (Fisher Scientific, Hampton, N.H.). For each subject, three sections, at intervals of approximately 300 μm, were selected for in situ hybridization.
The antisense riboprobe was transcribed in vitro from a 942-bp DNA fragment corresponding to bases 900–1841 within the protein coding region of the human KCNS3 gene (NM_002252) in the presence of 35S-CTP (PerkinElmer, Waltham, Mass.). The specificity of this riboprobe was demonstrated in our previous study (14). In situ hybridization procedures for KCNS3 mRNA were performed as previously described (14) (see the Supplemental Methods section of the online data supplement). The hybridized sections were exposed to autoradiographic films, coated with nuclear emulsion, developed, and counterstained with cresyl violet. Sections were processed in a pairwise manner throughout the in situ hybridization procedures and subsequent signal detections.
KCNS3 mRNA levels in the gray matter of prefrontal cortex area 9 were measured as film optical densities, as previously described (10) (see the Supplemental Methods section of the online data supplement). To evaluate KCNS3 mRNA expression at the cellular level, the numbers of silver grains generated by the 35S-labeled riboprobe in emulsion-coated sections were counted in sampling frames that were systematically and randomly placed in the gray matter of prefrontal cortex area 9, as previously described (5) (see Figure S1 in the data supplement). Within the sampling frame, grain numbers were counted within 22-μm-diameter circles placed over each Nissl-stained nucleus. Neurons were considered to be specifically labeled, referred to as KCNS3 mRNA-expressing neurons, if the grain number per neuron was at least five times the background number, as determined for each section by counting the grains within 22-μm-diameter circles placed over glial nuclei (5, 24) (see the Supplemental Methods section of the online data supplement).
Microarray Analysis
For laser microdissection of individual neurons, cryostat sections of prefrontal cortex area 9 were dual-labeled with lectin Vicia villosa agglutinin (VVA) and anti-NeuN antibody in order to visualize parvalbumin neuron-selective perineuronal nets (25) and all neurons, respectively (see the Supplemental Methods section of the online data supplement). Dual-labeled cells (parvalbumin neurons) and VVA-negative/NeuN-positive cells (non-parvalbumin neurons) were individually collected using laser microdissection from layers 3 and 4 for each subject. Pilot quantitative polymerase chain reaction studies (not shown) confirmed that levels of GAD67 and parvalbumin mRNAs in the dual-labeled neurons were 40.1 times and 68.5 times greater, respectively, than in VVA-negative/NeuN-positive cells.
For each subject of the 14 pairs, RNA was extracted from pooled samples of 360 VVA-labeled neurons, converted into cDNA, amplified, labeled with biotin, and loaded on Affymetrix GeneChip HT HG-U133+ PM Array Plate (Affymetrix, Santa Clara, Calif.). Scanned images were segmented and converted into DAT files, using Microarray Analysis Suite 5.0. Segmented images were normalized and log2-transformed using GeneChip Robust Multiarray Average (26).
Antipsychotic-Exposed Monkeys
Three groups (N=6 per group) of young adult male macaque monkeys (Macaca fascicularis) were exposed for 17–27 months to oral haloperidol, olanzapine, or placebo at dosages that produced trough plasma levels in the therapeutic range for the treatment of schizophrenia (27). Triads of monkeys, one from each of the three groups, were euthanized together on separate days. For each animal, two coronal sections of the right frontal lobe spaced 200 μm apart were hybridized with the antisense riboprobe against human KCNS3 mRNA. The film optical density was measured in the dorsal and ventral banks of the principal sulcus, corresponding to prefrontal cortex area 46. All animal studies followed the NIH Guide for the Care and Use of Laboratory Animals and were approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
Statistical Analysis
Statistical analyses were performed using SPSS (SPSS, Inc., Chicago). The significance threshold was set at 0.05. For each of three in situ hybridization measures of KCNS3 mRNA expression (film optical density, the density of KCNS3 mRNA-expressing neurons, and grain number per neuron), data were averaged across the three sections in each subject before statistical analyses. Analyses of covariance (ANCOVA) were first conducted to determine whether the in situ hybridization measures were related to sex, age, postmortem interval, brain pH, RIN, and storage time. We found that the film optical density significantly covaried with age (F=8.9, df=1, 36, p=0.005) and RIN (F=5.3, df=1, 36, p=0.03) and that the density of KCNS3 mRNA-expressing neurons significantly covaried with storage time (F=7.3, df=1, 36, p=0.01). No other factors were found to be correlated with any of the in situ hybridization measures of KCNS3 mRNA expression (Table 1, Table S1). Consequently, the ANCOVA models we report included each of the in situ hybridization measures as the independent variable, diagnostic group as the main effect, and subject pair as a blocking factor that reflects the matching of individual subject pairs for sex and age. In addition, we included as covariates in each model any subject characteristic that significantly covaried with the in situ hybridization measures but that were not used in the matching of subjects pairs (i.e., RIN for the film density and storage time for the density of KCNS3-expressing neurons). Although sex, age, and their interaction have robust impacts on global gene expression in the human cortex (28, 29), matching only these factors may not be sufficient to control for sources of biological variability. Thus, to validate the paired ANCOVA models, we performed unpaired ANCOVA models without subject pair as a blocking factor. In these models, age and RIN were added as covariates for the film optical density, and storage time was included as a covariate for the density of KCNS3-expressing neurons. The relationship between KCNS3 mRNA levels and age was assessed by Pearson’s correlation analysis.
In microarray analyses, normalized and log2-transformed signals were compared between schizophrenia and comparison subjects using the paired t test. The use of the paired t test reflects the pairing of schizophrenia and comparison subjects based on sex and age. For KCNS3 mRNA, the t test was one-tailed, because of the direction of difference predicted from the in situ hybridization findings in the first cohort. For KCNB1 mRNA, which encodes the Kv2.1 subunit, the differential expression between schizophrenia and comparison subjects was tested using a two-tailed paired t test. For both KCNS3 and KCNB1 mRNAs, we calculated log2-transformed signal ratios between schizophrenia and comparison subjects for each pair. Pearson’s correlation analysis was used to assess the relationship between KCNS3 and KCNB1 mRNA microarray signals across the 28 subjects as well as the relationship between the within-subject-pair schizophrenia-to-comparison subject signal ratios of these mRNAs across the 14 subject pairs.
The effects of antipsychotic exposure on KCNS3 mRNA expression levels in monkeys were evaluated by a single-factor analysis of variance model with treatment group as the main effect and triad as a blocking factor (10).
Results
KCNS3 mRNA Expression in Prefrontal Cortex Area 9 of Schizophrenia and Comparison Subjects
As previously reported, in comparison subjects, KCNS3 mRNA signals detected with 35S-labeled antisense riboprobe were high in layers 2 to 5, with the highest signal density in layer 4 (Figure 1A; see also Figure S1). Layer 6 exhibited the lowest signal density, and no specific hybridization signal was detected in layer 1 (Figure 1A; Figure S1). In schizophrenia subjects, KCNS3 mRNA expression appeared to be decreased across layers 2 to 5 relative to matched comparison subjects (Figure 1A). Quantification of film optical densities in the gray matter confirmed that mean KCNS3 mRNA levels were 23% lower in schizophrenia subjects than in comparison subjects (9.9 nCi/g [SD=6.9] compared with 12.9 nCi/g [SD=6.2]; paired ANCOVA: F=9.3, df=1, 20, p=0.006; unpaired ANCOVA: F=5.0, df=1, 40, p=0.03) (Figure 1B). The mRNA levels were lower in the schizophrenia subjects for 18 of the 22 subject pairs.
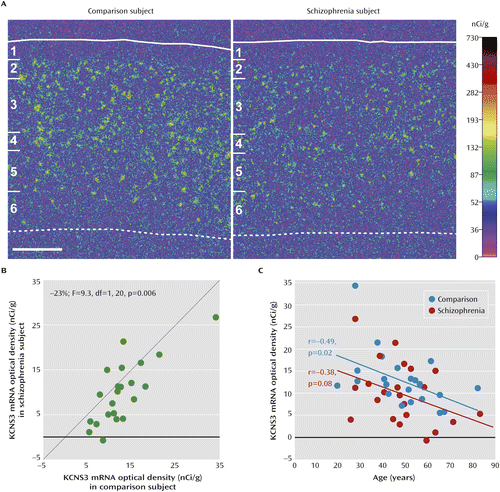
a In panel A, pseudocolored film autoradiographs of sections containing prefrontal cortex area 9 processed by in situ hybridization demonstrate lower KCNS3 mRNA levels in a schizophrenia subject relative to the matched comparison subject. The solid white line indicates the border between pia mater and layer 1, and the dotted line indicates the border between layer 6 and the white matter. The six cortical layers are identified on the left in each panel. Scale bar=1 mm. In panel B, average KCNS3 mRNA levels across gray matter of prefrontal cortex area 9 for schizophrenia subjects relative to matched comparison subjects are plotted for each pair. Green circles represent each subject pair. Data points to the right of the unity line indicate lower mRNA levels in the schizophrenia subject relative to the comparison subject and vice versa. Mean KCNS3 mRNA levels were 23% lower in schizophrenia subjects relative to matched comparison subjects. In panel C, KCNS3 mRNA expression levels in prefrontal cortex area 9 are plotted against age for schizophrenia subjects and comparison subjects. KCNS3 mRNA levels were significantly negatively correlated with age in comparison subjects, and the correlation fell short of significance in schizophrenia subjects. The regression line for schizophrenia subjects is parallel to and shifted downward from that for comparison subjects, suggesting that the decreased expression of KCNS3 mRNA is similar in magnitude across adult life.
Because we detected a significant covariate effect of age on the film optical density of KCNS3 mRNA levels, we performed correlation analyses. KCNS3 mRNA levels were negatively correlated with age in comparison subjects (r=−0.49, p=0.02); in schizophrenia subjects the correlation was also negative but fell short of statistical significance (r=−0.38, p=0.08) (Figure 1C).
Cellular Levels of KCNS3 mRNA Expression in Schizophrenia and Comparison Subjects
At the cellular level, silver grain clusters representing KCNS3 mRNA expression were detected over neuronal nuclei in the emulsion coated slides (Figure S1). The mean density of KCNS3 mRNA-expressing neurons was 32% lower in schizophrenia subjects relative to comparison subjects (18.4 neurons/mm2 [SD=8.2] compared with 27.2 neurons/mm2 [SD=6.8]; paired ANCOVA: F=30.9, df=1, 20, p<0.001; unpaired ANCOVA: F=20.7, df=1, 41, p<0.001) (Figure 2A). Furthermore, the mean number of grains per neuron was 16% lower in the schizophrenia subjects than in the comparison subjects (41.2 [SD=9.9] compared with 49.3 [SD=9.4]; paired ANCOVA: F=28.6, df=1, 21, p<0.001; unpaired ANCOVA: F=7.8, df=1, 42, p=0.008) (Figure 2B).
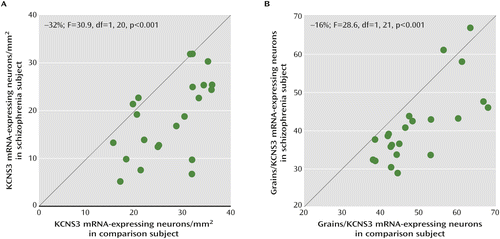
a Average numbers of KCNS3 mRNA-expressing neurons/mm2 (panel A) and average grain numbers per KCNS3 mRNA-expressing neuron (panel B) for schizophrenia subjects relative to matched comparison subjects are plotted for each pair. Green circles represent each subject pair. Data points to the right of the unity line indicate lower measures in the schizophrenia subject relative to the comparison subject and vice versa. In schizophrenia subjects, both the mean density of KCNS3 mRNA-expressing neurons and the mean grain number per KCNS3 mRNA-expressing neuron were lower (by 32% and 16%, respectively) relative to comparison subjects.
Microarray Analysis
In a separate cohort of 14 matched pairs of schizophrenia and comparison subjects, parvalbumin neurons were isolated by laser microdissection based on the selective labeling of their perineuronal nets with the plant lectin VVA (25) (Figure 3). Mean levels of KCNS3 mRNA in VVA-labeled neurons were 40% lower in schizophrenia subjects (t=1.97, p=0.04 with one-tailed paired t test) (Figure 3D). Interestingly, KCNB1 mRNA, which encodes Kv2.1 subunits that associate with KCNS3-encoded Kv9.3 subunits to form heteromeric channels, also displayed a mean decrease of 41% in schizophrenia subjects (Figure 3E), although this did not reach statistical significance (t=1.76, p=0.10 with two-tailed paired t test). Correlation analyses revealed a significant positive correlation (r=0.50, p=0.007) between KCNS3 and KCNB1 mRNA levels across all 28 subjects (Figure 3F) and a nonsignificant positive correlation in the within-subject-pair schizophrenia-to-comparison signal ratios between KCNS3 and KCNB1 mRNAs across the 14 subject pairs (r=0.48, p=0.09) (Figure 3G).
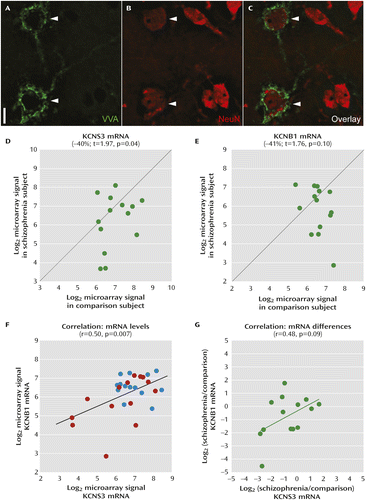
a Representative photomicrographs demonstrate dual-fluorescence labeling using biotinylated VVA to detect perineuronal nets (panel A) and an antibody against the neuronal protein NeuN (panel B). In panel C, an overlay of panels A and B illustrates that only some neurons (arrowheads) are surrounded by perineuronal nets. Scale bar=10 μm. Log2-transformed microarray signals of KCNS3 (panel D) and KCNB1 (panel E) mRNAs for schizophrenia subjects relative to matched comparison subjects are plotted for each pair. Green circles represent each subject pair. Data points to the right of the unity line indicate lower mRNA signals in the schizophrenia subject relative to the comparison subject and vice versa. Mean KCNS3 and KCNB1 mRNA levels were decreased by 40% and 41%, respectively, in schizophrenia subjects relative to matched comparison subjects. In panel F, log2-transformed microarray signals for KCNS3 and KCNB1 mRNAs are plotted across 28 subjects, with blue and red circles representing comparison and schizophrenia subjects, respectively. Pearson’s correlation analysis revealed a significant correlation between KCNS3 and KCNB1 mRNA levels. In panel G, log2-tranformed signal ratios between schizophrenia and comparison subjects were calculated for both KCNS3 and KCNB1 mRNAs in each pair and plotted across 14 pairs. Green circles represent each subject pair. Pearson’s correlation analysis detected a trend toward correlated reductions of KCNS3 and KCNB1 mRNAs in schizophrenia.
Relationship Between Clinical Factors and Lower KCNS3 mRNA Expression Levels in Schizophrenia
We assessed whether the differences in KCNS3 mRNA levels between schizophrenia and comparison subjects were influenced by clinical factors in schizophrenia subjects, such as sex, family history, age at illness onset, suicide as the cause of death, history of marriage, socioeconomic status, independence of living at time of death, presence of any substance abuse or dependence at time of death, and use of antidepressants, benzodiazepines/anticonvulsants, or antipsychotics at time of death (see the Supplemental Methods section of the online data supplement). None of these factors had a significant effect on the within-subject pair differences as determined by in situ hybridization film optical densities or microarray signals (see Figure S2 in the data supplement).
To test whether chronic exposure to conventional or atypical antipsychotics could affect KCNS3 mRNA expression, we examined optical densities of the hybridization signals in the prefrontal cortex of monkeys chronically exposed to haloperidol, olanzapine, or placebo. Mean KCNS3 mRNA levels did not significantly differ between the three groups (placebo: 13.7 nCi/g [SD=2.1]; haloperidol: 14.1 nCi/g [SD=2.9]; olanzapine: 13.0 nCi/g [SD=2.0]) (see Figure S3 in the data supplement).
Discussion
In schizophrenia, previous postmortem studies revealed various abnormalities in cortical GABA neurons, including expression deficits of genes that regulate GABA neurotransmission (9, 23, 30). Some of these changes are prominent in parvalbumin neurons and are thought to underlie disturbances in γ-oscillations in schizophrenia (3, 13). In this study, we focused on the parvalbumin neuron-selective KCNS3 voltage-gated potassium channel subunit gene that appears to have a physiological function in the generation of γ-oscillations by parvalbumin neurons. Our in situ hybridization analyses of 22 pairs of schizophrenia and matched comparison subjects revealed that the density of cortical KCNS3 mRNA-expressing neurons and the mean KCNS3 mRNA level per neuron were reduced in the schizophrenia subjects, indicating that KCNS3 mRNA levels are decreased in the majority of parvalbumin neurons and below the threshold of detection in a subset of severely affected parvalbumin neurons. Consistently, microarray analyses specifically of prefrontal cortex neurons labeled with VVA, a selective marker of parvalbumin neurons, in a separate cohort of 14 subject pairs detected significantly lower KCNS3 mRNA levels in schizophrenia subjects. In addition to previous findings in parvalbumin neurons, such as lower GAD67 expression, that appear to represent a molecular basis for reduced magnitude of GABA neurotransmission by these neurons (3), the lower KCNS3 mRNA levels in prefrontal cortical parvalbumin neurons, demonstrated by two different methods in two nonoverlapping cohorts, could provide another molecular mechanism of parvalbumin neuron dysfunction that might also underlie the disturbances in γ-oscillations in schizophrenia.
Consistent with the reduced density of cortical perineuronal nets in schizophrenia (31), VVA labeling appeared to be lower in the schizophrenia subjects. This observation raises the possibility that some perineuronal nets were not visible, and thus the corresponding parvalbumin neurons were not sampled in schizophrenia subjects. However, a reduction in VVA labeling in schizophrenia would likely bias against finding a difference in KCNS3 expression in schizophrenia, as the most affected cells would be the least likely to be captured and assayed.
Several lines of evidence indicate that the reduction in KCNS3 mRNA expression is due to the disease process of schizophrenia and not attributable to potential confounding factors frequently associated with the illness. First, among the 22 schizophrenia subjects analyzed by in situ hybridization, four subjects who were not taking antipsychotic medications at time of death demonstrated lower KCNS3 mRNA levels relative to their matched comparison subjects, similar to the lower levels seen in 18 schizophrenia subjects on medication at time of death (Figure S2). Second, KCNS3 mRNA expression was unaltered in the prefrontal cortex of monkeys chronically exposed to either haloperidol or olanzapine (Figure S3). Third, none of the potentially confounding clinical factors, such as suicide, socioeconomic status, substance abuse or dependence at time of death, and treatment with antidepressants or benzodiazepines/anticonvulsants at time of death, accounted for the lower KCNS3 mRNA expression in the gray matter or individual VVA-labeled neurons in schizophrenia subjects (Figure S2).
A significant negative correlation between age and the KCNS3 mRNA expression was observed in comparison subjects, and the correlation fell short of significance in schizophrenia subjects. The regression line for schizophrenia subjects was parallel to and shifted downward from that of comparison subjects (Figure 1C), indicating that the decreased expression of KCNS3 mRNA in schizophrenia is present across adult life and thus unlikely to be a consequence of illness chronicity. Furthermore, this observation suggests that the KCNS3 expression deficit may be present early in the course of the illness and thus could contribute to the pathophysiology of the clinical features of the illness.
KCNS3 mRNA encodes potassium channel Kv9.3 modulatory α-subunits. Kv9.3 subunits assemble with ubiquitously expressed Kv2.1 α-subunits, with a 1:3 subunit stoichiometry, to form heteromeric Kv2.1/Kv9.3 channels (17). Interestingly, in the microarray data of individually collected VVA-labeled neurons, KCNB1 mRNA, which encodes the Kv2.1 subunit, showed a decrease comparable to KCNS3 mRNA in schizophrenia (Figure 3E), and expression levels of both transcripts tracked together (Figure 3F, 3G). These findings suggest a concomitant down-regulation of Kv9.3 and Kv2.1 subunits, and thus a lower complement of Kv2.1/Kv9.3 heteromeric channels, in prefrontal cortical parvalbumin neurons in schizophrenia (Figure 4).

a In panel A, a schematic diagram illustrates a parvalbumin neuron (PV) that receives excitatory synapses from, and makes perisomatic inhibitory synapses on, neighboring pyramidal neurons (I–IV). In panel B, in unaffected subjects (left column), normal expression of Kv2.1/Kv9.3 heteromeric channels contributes to a normal level of assembled potassium channels in the cell membrane, which leads to fast excitatory postsynaptic potentials (EPSPs) in response to the firing of pyramidal neurons (I–IV). Because fast EPSPs summate during a short time window, the parvalbumin neuron fires only when it receives coincident excitatory inputs (i.e., EPSPs) from these pyramidal neurons (RMP=resting membrane potential). This ability of parvalbumin neurons to detect the synchronous firing of multiple pyramidal neurons, together with their perisomatic innervation of these pyramidal neurons, appears to be critical to the generation of γ-oscillations. In schizophrenia subjects (right column), lower levels of Kv2.1/Kv9.3 channels result in slower EPSPs, and thus even noncoincident EPSPs can sum to trigger action potentials in parvalbumin neurons. This alteration would compromise the detection of coincident EPSPs by parvalbumin neurons and impair the generation of γ-oscillations.
The subcellular distribution of Kv2.1 immunoreactivity in cortical parvalbumin neurons supports the presence of Kv2.1/Kv9.3 heteromeric channels in the dendrites of individual parvalbumin neurons (18). Compared with homomeric Kv2.1 channels, Kv2.1/Kv9.3 channels are more effectively activated by small depolarization steps from the resting membrane potential (15, 16). These data suggest that Kv2.1/Kv9.3 channels are among those dendritic voltage-gated potassium channels that contribute to the acceleration of excitatory postsynaptic potential decay in parvalbumin neurons (22). Because such fast potentials summate in a narrow time window (22), parvalbumin neurons can efficiently detect temporally convergent excitatory inputs and fire action potentials (Figure 4). The output of parvalbumin neurons simultaneously inhibits large groups of neighboring pyramidal neurons (32, 33), leading to their synchronized firing when they concomitantly emerge from the hyperpolarized state (34). This synchrony occurs as a γ-frequency oscillation because of the rate of decay of the pyramidal cell inhibition at synapses from parvalbumin neurons (34). Because parvalbumin neurons receive excitatory inputs from neighboring pyramidal neurons (35), the presence of Kv2.1/Kv9.3 channels in parvalbumin neurons enables them to fire action potentials in response to the coincident inputs from the synchronized activity of pyramidal neurons, sustaining γ-oscillations in the parvalbumin-pyramidal neuron network. Consequently, the deficient KCNS3 expression, and the concomitant down-regulation of KCNB1 expression in parvalbumin neurons, would be predicted to slow the time course of excitatory postsynaptic potentials in parvalbumin neurons, contributing to altered γ-oscillations (Figure 4) and impaired cognitive function in schizophrenia (13).
Abnormal cortical expression has been reported for other voltage-gated potassium channel subunit genes—namely, KCNC1 (Kv3.1) (36) and KCNH2 (Kv11.1) (37)—in schizophrenia, although the affected cell types were not determined. In contrast to the KCNS3-encoded Kv9.3 subunit that controls subthreshold membrane potentials (15), these subunits regulate repolarization of action potentials (37, 38), a process critical to the high-frequency and repetitive firing of parvalbumin neurons. Therefore, alterations in different types of potassium channel subunits may disturb different electrophysiological properties of parvalbumin neurons, which could converge on disturbances in γ-oscillations in schizophrenia. Because KCNS3 is selectively expressed in parvalbumin neurons, the identification of pharmacological compounds targeting Kv9.3 subunits may be useful for developing therapeutic strategies that selectively restore the function of parvalbumin neurons, enhance cortical synchronization, and improve cognition in schizophrenia.
1 : The neurobiology of cognition in schizophrenia. J Clin Psychiatry 2006; 67:e11Crossref, Medline, Google Scholar
2 : Circuit-based framework for understanding neurotransmitter and risk gene interactions in schizophrenia. Trends Neurosci 2008; 31:234–242Crossref, Medline, Google Scholar
3 : Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends Neurosci 2012; 35:57–67Crossref, Medline, Google Scholar
4 : Interneuron dysfunction in psychiatric disorders. Nat Rev Neurosci 2012; 13:107–120Crossref, Medline, Google Scholar
5 : Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J Neurosci 2003; 23:6315–6326Crossref, Medline, Google Scholar
6 : Cortical deficits of glutamic acid decarboxylase 67 expression in schizophrenia: clinical, protein, and cell type-specific features. Am J Psychiatry 2011; 168:921–929Link, Google Scholar
7 : Alterations in chandelier neuron axon terminals in the prefrontal cortex of schizophrenic subjects. Am J Psychiatry 1999; 156:1709–1719Abstract, Google Scholar
8 : Molecular determinants of dysregulated GABAergic gene expression in the prefrontal cortex of subjects with schizophrenia. Biol Psychiatry 2009; 65:1006–1014Crossref, Medline, Google Scholar
9 : Expression of interneuron markers in the dorsolateral prefrontal cortex of the developing human and in schizophrenia. Am J Psychiatry 2010; 167:1479–1488Link, Google Scholar
10 : Deficits in transcriptional regulators of cortical parvalbumin neurons in schizophrenia. Am J Psychiatry 2012; 169:1082–1091Link, Google Scholar
11 : Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature 2009; 459:663–667Crossref, Medline, Google Scholar
12 : Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature 2009; 459:698–702Crossref, Medline, Google Scholar
13 : Abnormal neural oscillations and synchrony in schizophrenia. Nat Rev Neurosci 2010; 11:100–113Crossref, Medline, Google Scholar
14 : Selective expression of KCNS3 potassium channel α-subunit in parvalbumin-containing GABA neurons in the human prefrontal cortex. PLoS ONE 2012; 7:e43904Crossref, Medline, Google Scholar
15 : Kv2.1/Kv9.3, a novel ATP-dependent delayed-rectifier K+ channel in oxygen-sensitive pulmonary artery myocytes. EMBO J 1997; 16:6615–6625Crossref, Medline, Google Scholar
16 : Heteromeric assembly of Kv2.1 with Kv9.3: effect on the state dependence of inactivation. Biophys J 1999; 77:248–257Crossref, Medline, Google Scholar
17 : Fluorescence measurements reveal stoichiometry of K+ channels formed by modulatory and delayed rectifier alpha-subunits. Proc Natl Acad Sci USA 2005; 102:6160–6165Crossref, Medline, Google Scholar
18 : The K+ channel, Kv2.1, is apposed to astrocytic processes and is associated with inhibitory postsynaptic membranes in hippocampal and cortical principal neurons and inhibitory interneurons. Neuroscience 1998; 84:37–48Crossref, Medline, Google Scholar
19 : Synaptic efficacy during repetitive activation of excitatory inputs in primate dorsolateral prefrontal cortex. Cereb Cortex 2004; 14:530–542Crossref, Medline, Google Scholar
20 : Interneuron Diversity series: fast in, fast out: temporal and spatial signal processing in hippocampal interneurons. Trends Neurosci 2004; 27:30–40Crossref, Medline, Google Scholar
21 : Properties of excitatory synaptic responses in fast-spiking interneurons and pyramidal cells from monkey and rat prefrontal cortex. Cereb Cortex 2006; 16:541–552Crossref, Medline, Google Scholar
22 : Dendritic mechanisms underlying rapid synaptic activation of fast-spiking hippocampal interneurons. Science 2010; 327:52–58Crossref, Medline, Google Scholar
23 : Decreased glutamic acid decarboxylase67 messenger RNA expression in a subset of prefrontal cortical gamma-aminobutyric acid neurons in subjects with schizophrenia. Arch Gen Psychiatry 2000; 57:237–245Crossref, Medline, Google Scholar
24 : Dopamine differentially regulates dynorphin, substance P, and enkephalin expression in striatal neurons: in situ hybridization histochemical analysis. J Neurosci 1991; 11:1016–1031Crossref, Medline, Google Scholar
25 : Neocortical neuronal diversity: chemical heterogeneity revealed by colocalization studies of classic neurotransmitters, neuropeptides, calcium-binding proteins, and cell surface molecules. Cereb Cortex 1993; 3:273–289Crossref, Medline, Google Scholar
26 : Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5:R80Crossref, Medline, Google Scholar
27 : The influence of chronic exposure to antipsychotic medications on brain size before and after tissue fixation: a comparison of haloperidol and olanzapine in macaque monkeys. Neuropsychopharmacology 2005; 30:1649–1661Crossref, Medline, Google Scholar
28 : Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci USA 2008; 105:15605–15610Crossref, Medline, Google Scholar
29 : Spatio-temporal transcriptome of the human brain. Nature 2011; 478:483–489Crossref, Medline, Google Scholar
30 : Expression of GABA signaling molecules KCC2, NKCC1, and GAD1 in cortical development and schizophrenia. J Neurosci 2011; 31:11088–11095Crossref, Medline, Google Scholar
31 : Extracellular matrix-glial abnormalities in the amygdala and entorhinal cortex of subjects diagnosed with schizophrenia. Arch Gen Psychiatry 2010; 67:155–166Crossref, Medline, Google Scholar
32 : Hippocampal CA1 interneurons: an in vivo intracellular labeling study. J Neurosci 1995; 15:6651–6665Crossref, Medline, Google Scholar
33 : Synchronization of neuronal activity in hippocampus by individual GABAergic interneurons. Nature 1995; 378:75–78Crossref, Medline, Google Scholar
34 : GABA neurons and the mechanisms of network oscillations: implications for understanding cortical dysfunction in schizophrenia. Schizophr Bull 2008; 34:944–961Crossref, Medline, Google Scholar
35 : Pyramidal neuron local axon terminals in monkey prefrontal cortex: differential targeting of subclasses of GABA neurons. Cereb Cortex 2003; 13:452–460Crossref, Medline, Google Scholar
36 : Kv3.1-containing K+ channels are reduced in untreated schizophrenia and normalized with antipsychotic drugs. Mol Psychiatry (Epub ahead of print, April 30, 2013)Google Scholar
37 : A primate-specific, brain isoform of KCNH2 affects cortical physiology, cognition, neuronal repolarization and risk of schizophrenia. Nat Med 2009; 15:509–518Crossref, Medline, Google Scholar
38 : Kv3 channels: voltage-gated K+ channels designed for high-frequency repetitive firing. Trends Neurosci 2001; 24:517–526Crossref, Medline, Google Scholar



