The Endophenotype Concept in Psychiatry: Etymology and Strategic Intentions
Abstract
Endophenotypes, measurable components unseen by the unaided eye along the pathway between disease and distal genotype, have emerged as an important concept in the study of complex neuropsychiatric diseases. An endophenotype may be neurophysiological, biochemical, endocrinological, neuroanatomical, cognitive, or neuropsychological (including configured self-report data) in nature. Endophenotypes represent simpler clues to genetic underpinnings than the disease syndrome itself, promoting the view that psychiatric diagnoses can be decomposed or deconstructed, which can result in more straightforward—and successful—genetic analysis. However, to be most useful, endophenotypes for psychiatric disorders must meet certain criteria, including association with a candidate gene or gene region, heritability that is inferred from relative risk for the disorder in relatives, and disease association parameters. In addition to furthering genetic analysis, endophenotypes can clarify classification and diagnosis and foster the development of animal models. The authors discuss the etymology and strategy behind the use of endophenotypes in neuropsychiatric research and, more generally, in research on other diseases with complex genetics.
As we celebrate the 50th anniversary of Nobelists Watson, Crick, and Wilkin’s discovery (with Franklin) of the structure of DNA—and its offspring, the complete sequencing of the human genome—it is salutary to contemplate the relative youthfulness of the field of human genetics. The term “genetics” was provided by William Bateson in 1902 (the Wright brothers’ first flight was in 1903). In 1909, the clarifying distinction we now take for granted between the concept of “genotype” and the concept of “phenotype” was provided by the Danish botanist Wilhelm Johanssen. He also introduced the word “gene.” His research on self-fertilized lines of beans revealed that quantitative variability in the phenotype confounded thinking about separable contributions of heredity and environment. He found that the phenotype is often an imperfect indicator of the genotype, that the same genotype may give rise to a wide range of phenotypes, and that the same phenotype may have arisen from different genotypes. Specific evidence for multifactorial (genetic and nongenetic) contributions to a continuous phenotype was provided about the same time by H. Nilsson-Ehle on the basis of observations of seed colors in crosses of oats and wheat. However, the term “polygene” was not available until K. Mather coined it in 1941. Exact citations for these historical references, often in German, are provided in the classic text by A.H. Sturtevant (1).
Genotypes, which can be measured with techniques of molecular biology such as polymerase chain reaction (PCR) and DNA sequencing, are often useful as probabilistic prognosticators of disease. In contrast, a phenotype represents observable characteristics of an organism, which are the joint product of both genotypic and environmental influences. In diseases with classic or Mendelian genetics as their distal causes, genotypes are usually indicative of phenotypes. However, this degree of genetic certainty does not exist for diseases with complex genetics (2–4). Genetic probabilism aptly describes the process by which a particular genotype gives rise to phenotype (5, 6). Epigenetic factors may also be of critical importance for modifying the development of phenotypes (7), and such modifications may be influenced by genotype or environment or be entirely stochastic in origin (8). Thus, models of complex genetic disorders predict a ballet choreographed interactively over time among genotype, environment, and epigenetic factors, which gives rise to a particular phenotype (9–12).
Despite the successful characterization of the nucleotide base-pair order that represents the human genome (13, 14), and although a legion of genetic linkage and association studies have been done, psychiatry has had little success in definitively identifying “culprit” genes or gene regions in the development of diseases categorized by using the field’s diagnostic classification schemas (15–18). The reason there is so much difficulty is undoubtedly—in part—that psychiatry’s classification systems describe heterogeneous disorders (19–22). In addition to the inherent complexity of psychiatric diseases, which have multifactorial and polygenic origins, the brain is the most complex of all organs. In organs such as the liver, all cells are nearly identical in their phenotypes and very similar in their transcriptomes (mRNA transcripts) and proteomes. In addition to the homogeneity in the structure of such cells, their interactions are mostly homogeneous. However, individual cells of the brain are quite different from each other in their transcriptomes, proteomes, and morphological phenotypes and also in the thousands of connections and interactions with other neurons and glia that are critically important to optimal functioning. Different cellular experiences are transduced to differences on the biochemical and epigenetic levels so that cellular memories regulated by protein modification, morphometric changes, and epigenetic influences make the brain unique among organs. Furthermore, the brain is subject to complex interactions not just among genes, proteins, cells, and circuits of cells but also between individuals and their changing experiences (23). Therefore, the phenotypic output from the brain, i.e., behavior, is not simply a sum of all its parts. It stands to reason that more optimally reduced measures of neuropsychiatric functioning should be more useful than behavioral “macros” in studies pursuing the biological and genetic components of psychiatric disorders.
The Endophenotype Concept in Psychiatry
The theory that genes and environment combine to confer susceptibility to the development of diseases surfaced in the early half of the last century, but the use of such a framework for exploring the etiology of schizophrenia and other psychiatric disorders is more recent. Douglas Falconer’s 1965 multifactorial threshold model for diabetes and other common, non-Mendelizing diseases was adapted to a polygenic model of schizophrenia in 1967 (24). About this time, it became clear that the classification of psychiatric diseases on the basis of overt phenotypes (syndromic behaviors) might not be optimal for genetic dissection of these diseases, which have complex genetic underpinnings. In their writings summarizing genetic theories in schizophrenia 30 years ago, Gottesman and Shields (25, 26) described “endophenotypes” as internal phenotypes discoverable by a “biochemical test or microscopic examination.” The term was adapted from a 1966 paper by John and Lewis (27), who had used it to explain concepts in evolution and insect biology. They wrote that the geographical distribution of grasshoppers was a function of some feature not apparent in their “exophenotypes”; this feature was “the endophenotype, not the obvious and external but the microscopic and internal.”
That felicitous term seemed to suit the needs of psychiatric genetics, and the concept of endophenotype was adapted for filling the gap between available descriptors and between the gene and the elusive disease processes. The identification of endophenotypes, which do not depend on what was obvious to the unaided eye, could help to resolve questions about etiological models. The rationale for the use of endophenotypes in exploring disease processes is illustrated in Figure 1. This rationale held that if the phenotypes associated with a disorder are very specialized and represent relatively straightforward and putatively more elementary phenomena (as opposed to behavioral macros), the number of genes required to produce variations in these traits may be fewer than those involved in producing a psychiatric diagnostic entity. Endophenotypes provided a means for identifying the “downstream” traits or facets of clinical phenotypes, as well as the “upstream” consequences of genes and, in principle, could assist in the identification of aberrant genes in the hypothesized polygenic systems conferring vulnerabilities to disorders. That is, the intervening variables or hypothetical constructs that were championed as useful for theorizing about behaviors (35)—and that could mark the path between the genotype and the behavior of interest (Figure 2)—might Mendelize in a predicted manner.
Despite the inherent advantages of the concept of endophenotype, the term and its promise lay dormant for a number of years. However, now that multiple genetic linkage and association studies using current classification systems and the development of practical animal models, have all fallen short of success, the term and its usefulness have reemerged. (A MEDLINE search for the years 2000 through 2002 found 62 entries for “endophenotype,” compared with 16 entries before 2000.) Endophenotypes are being seen as a viable and perhaps necessary mechanism for overcoming the barriers to progress (28, 51–58). The methods available for endophenotype analysis have advanced considerably since 1972; our current armamentarium includes neurophysiological, biochemical, endocrinological, neuroanatomical, cognitive, and neuropsychological (including configured self-report data) measures (29). Advanced tools of neuroimaging such as functional magnetic resonance imaging (fMRI), morphometric MRI, diffusion tensor imaging, single photon emission computed tomography (SPECT), and positron emission tomography (PET) promise to expand the possibilities even more (30, 59–61). Other terms with patently synonymous meaning, such as “intermediate phenotype,” “biological marker,” “subclinical trait,” and “vulnerability marker,” have been used interchangeably. These terms may not necessarily reflect genetic underpinnings but may rather reflect associated findings (see the discussion in the next section). In this context, we use the term “biological marker” to signify differences that do not have genetic underpinnings and “endophenotype” when certain heritability indicators are fulfilled.
Endophenotypes in Genetic Analysis
An endophenotype-based approach has the potential to assist in the genetic dissection of psychiatric diseases. Endophenotypes would ideally have monogenic roots; however, it is likely that many would have polygenic bases themselves. Furthermore, the use of endophenotypes in genetic research must be tempered by the realization that without controls and limits, their usefulness may be obscured. For example, putative endophenotypes do not necessarily reflect genetic effects. Indeed, these biological markers may be environmental, epigenetic, or multifactorial in origin. Criteria useful for the identification of markers in psychiatric genetics have been suggested (62) and have been adapted here to apply to endophenotypes:
1. The endophenotype is associated with illness in the population.
2. The endophenotype is heritable.
3. The endophenotype is primarily state-independent (manifests in an individual whether or not illness is active).
4. Within families, endophenotype and illness co-segregate.
Subsequently, an additional criterion that may be useful for identifying endophenotypes of diseases that display complex inheritance patterns was suggested (29):
5. The endophenotype found in affected family members is found in nonaffected family members at a higher rate than in the general population.
Other fields of medicine have had some success in using endophenotypes to assist with genetic linkage studies. For instance, the multiple genes that cause long QT syndrome were identified by using an endophenotype-based method (63, 64). Manifestations of long QT syndrome include syncope, ventricle arrhythmias, and sudden death (63). Although not all family members who carry the disease genes show these symptoms, a much greater percentage have QT elongation as measured by ECG. By using QT elongation as a phenotype—and excluding or including pedigree members with this finding—linkage studies were successful in identifying the genes that cause the QT elongation endophenotype and thus the syndrome phenotypes of syncope, ventricle arrhythmias, and sudden death (64, 65). The identification of these genes has allowed for genetic manipulations in mice to study disease pathology and to further the development of novel medications (66). Other examples in the literature of endophenotype-based strategies for identifying genetic linkage include studies of idiopathic hemochromatosis (excessive serum iron) (67), juvenile myoclonic epilepsy (an EEG abnormality) (68), and familial adenomatous polyposis coli (intestinal polyps) (69). In other disorders with complex genetics such as diabetes, hypercholesterolemia, or hypertension, researchers use physiological challenges, biochemical assays, and physiological measures to obtain a primary index of disease pathology. Indeed, these syndromes may all present to the physician as fatigue, but the pathophysiological underpinnings are substantially different. The glucose tolerance test, measurements of serum cholesterol levels, and sphygmomanometer measurements all represent objective, quantifiable methods for making disease diagnosis and classification. In addition to being crucial in diagnosis and classification of these diseases, the phenomena measured by these methods constitute endophenotypes that represent the primary inclusion/exclusion feature by which “hits” for genetic linkage and association studies are defined.
In psychiatry, a number of attempts have been made to develop and determine the feasibility of candidate endophenotypes. However, few have met all the criteria listed earlier. Nonetheless, some linkage and association studies—using endophenotypes—have had moderate success. Candidate endophenotypes have also been used in the development of animal models and to subtype patients for classification and diagnostic reasons (see the discussion in later sections). The hunt for candidate endophenotypes has been described in the literature on several psychiatric disorders, including schizophrenia (30, 31–33, 39, 70–73), mood disorders (28, 55, 74, 75), Alzheimer’s disease (76, 77), attention deficit hyperactivity disorder (54, 78, 79), and even personality disorders (80). We give a brief description of some possibilities in schizophrenia research as salient examples. The interested reader is referred to the references just cited for more in-depth discussions.
Sensory Motor Gating and Eye-Tracking Dysfunction in Schizophrenia
Deficits in sensory motor gating are consistent neuropsychological findings in schizophrenia (33, 39). The hypothesized association between these deficits and schizophrenia has face validity primarily on the basis of patients’ reports that they have difficulty filtering information from multiple sources (33, 81–83). On the level of neurobiology, the inhibitory mechanisms of patients with schizophrenia may not be capable of adequately adjusting to the multiple distinct or repetitive inputs that occur in everyday life. Neuropsychological tests, including assessments of P50 suppression and prepulse inhibition of the startle response, have been developed to discern efficiencies in these capabilities. Both tasks have been studied in schizophrenic patients, and abnormalities consistent with defects in inhibitory neuronal circuits have been found.
In tests of prepulse inhibition, startling sensory stimuli (loud noise, bright light) are used to elicit an unconditional reflexive startle response in individuals. If a weaker prestimulus is provided before the startling stimulus, the subsequent startle response is generally diminished. A relatively reproducible finding is that this dimunition of the second response is attenuated in patients with schizophrenia, compared to healthy subjects (39, 84, 85). Prepulse inhibition is a generally conserved finding among vertebrates, and as such it has been the target of several rodent studies (reviewed in reference 86), both to model a facet of schizophrenia and to investigate the biology of a prepulse inhibition response. The presence of this candidate endophenotype has been documented in relatives of patients with schizophrenia (87), but more extensive testing is required. Genetic studies in inbred animals have suggested at least a partial genetic diathesis (86); however, environmental influences may also be active (88, 89). Abnormal prepulse inhibition is not specific to schizophrenia; studies have identified this abnormality in obsessive-compulsive disorder (90) and Huntington’s disease (91), among others. However, the reproducibility of the finding in schizophrenia, the fact that abnormal prepulse inhibition parallels a putative central abnormality in the disease, and the fact that prepulse inhibition is a conserved phenomenon among vertebrates make abnormal prepulse inhibition a promising candidate endophenotype to pursue.
The P50 suppression test uses two auditory stimuli presented at 500-msec intervals. A positive event-related response for both stimuli is measured by EEG. In normal individuals, the neuronal response to the second stimulus is of lower amplitude than the first. However, patients with schizophrenia do not show the same degree of suppression of P50 amplitude (33, 92–95). In addition to this finding in probands, abnormal P50 suppression is found in unaffected first-degree relatives of patients with schizophrenia (95–99). The heritability of this measure has been assessed in twins, and the results have suggested that genetics plays a role in the development of variation in this candidate endophenotype (100, 101). Freedman and colleagues (102) also used P50 suppression to identify a potential susceptibility locus for schizophrenia on chromosome 15, a chromosomal region where the gene for the α7 nicotinic acetylcholine receptor resides. Furthermore, this group of researchers has shown linkage disequilibrium in this region (103) and has shown that promoter variants of the α7 receptor are associated with schizophrenia and/or P50 suppression abnormalities (104).
Eye-tracking dysfunction has long been associated with schizophrenia. This dysfunction was first described in 1908 by Diefendorf and Dodge (105), whose work was rediscovered in the 1970s, initially by Holzman and colleagues (106, 107).
Eye movements are generally of two forms, either saccadic (brief and extremely rapid movements) or smooth and controlled. The latter “smooth pursuit” eye movements occur only when the subject is following an object moving at a constant velocity, most commonly a pendulum (in early studies) or bright dot on a computer monitor. Initiation and maintenance of smooth pursuit eye movements involve integration of functions of the prefrontal cortex frontal eye fields, visual and vestibular circuitry, thalamus, and cerebellum, as well as the muscles and neural circuitry directly responsible for eye movement (108).
A number of studies have found that patients with schizophrenia have deficiencies in smooth pursuit eye movements, compared to healthy subjects (see references 41–43 for review). In general, these deficiencies are manifested as corrective saccades, which follow smooth pursuit eye movements that are slightly slower than the target (reviewed in reference 42, where more detailed descriptions of specific abnormalities are available). Furthermore, the heritability of these deficiencies has been extensively addressed; studies have suggested that biological relatives of schizophrenic subjects have an increased rate of smooth pursuit eye movement dysfunction. Thus, 40%–80% of schizophrenic subjects, 25%–45% of their first-degree relatives, and less than 10% of healthy comparison subjects generally show this trait (41–43). A study requiring replication has suggested linkage to a region of chromosome 6 (109). Correlating smooth pursuit function with neuroimaging measures (110) or performance on working memory tasks (111, 112) may be a useful research strategy. Smooth pursuit eye movements are maintained in primates but not in most other mammals used in preclinical research (108).
Working Memory in Schizophrenia
Working memory and executive cognition are compromised in patients with schizophrenia (44). A primary brain region involved in working memory is the dorsolateral prefrontal cortex (31, 45, 113), a region in which abnormalities have been found in postmortem studies of schizophrenic patients (114). Family (115, 116), and twin studies (117, 118) have suggested heritability of working memory deficits in schizophrenia.
Recent studies have identified gene and chromosomal regions possibly involved in working memory. A study of Finnish twins by Gasperoni and colleagues (53), which used an endophenotype-based strategy, suggested linkage and association to a region of chromosome 1. In their study, dizygotic twins discordant for schizophrenia underwent four neuropsychological tests. Using the sum of performance scores on these tests, Gasperoni and colleagues identified significant linkage to 1q41, a region previously suggested in traditional linkage studies of schizophrenia (119–122). By stratifying their data according to performance on each neuropsychological test, they found that visual working memory performance was highly significantly linked with this region (p=0.007), while performance on none of the other three neuropsychological tests was significantly associated with any 1q markers. In the second part of their study, Gasperoni and colleagues (53) completed an association analysis involving monozygotic discordant twins, unaffected dizygotic and monozygotic twins, and the dizygotic twin group from the linkage study. In this analysis, an association of the 1q41 region and performance on the visual working memory task was again identified. The facts that previous linkage studies have identified this region and that performance on working memory tasks is a reproducible endophenotype for schizophrenia strengthen the claim that this endophenotype—and the putative gene(s) at 1q41 linked to it—may be relevant to the pathophysiology of schizophrenia. The study requires replication in a larger group of subjects representing a nonisolate population.
Association and physiological evidence have also linked a specific enzyme with a small increased risk for developing schizophrenia and with poorer performance on a working memory task. The enzyme catechol O-methyltransferase (COMT), the gene for which is found at 22q11.2, assists in the catabolism of dopamine. This chromosomal region has been linked to both schizophrenia and bipolar disorder and overlaps with a deletion that has been associated with velocardiofacial syndrome (DiGeorge syndrome) and schizophrenia (see reference 16 for review). A functional polymorphism (val108/158met) for COMT results in a fourfold increase in the activity of this enzyme. The considerable body of evidence implicating dopaminergic neurotransmission, the presence of a common functional polymorphism, and the data suggesting the involvement of the dorsolateral prefrontal cortex in schizophrenia and working memory led to association studies of COMT (31).
While their effect sizes are small, a number of family studies have found that the valine allele is transmitted at a higher rate than the methionine allele to patients with schizophrenia than to their nonaffected siblings (reviewed in reference 31). This polymorphism has also been linked to performance on a working memory task. Specifically, Egan et al. (123) associated poorer performance on a working memory task in patients, their siblings, and comparison subjects with the same valine allele variation of COMT found to be transmitted at a higher rate in schizophrenia. They used fMRI to measure dorsolateral prefrontal cortex activation in a subset of these individuals; the fMRI fingerprint from individuals with the valine allele suggested that activation of the dorsolateral prefrontal cortex is less efficient in those subjects (123). Additional studies from two independent laboratories have also suggested that patients with schizophrenia show this inefficiency (124–126). Callicott and colleagues (127) have recently shown that the fMRI response in the dorsolateral prefrontal cortex observed in schizophrenic subjects is also found in unaffected siblings of patients with schizophrenia. Although they found no group differences between the siblings of schizophrenic patients and the comparison group in overall working memory performance, fMRI measurement showed that the sibling group had less efficient dorsolateral prefrontal cortex functioning than the comparison group. Taken together, these results suggest that fMRI analysis of subjects undergoing working memory tasks may be a more sensitive endophenotype than working memory performance alone as measured by neuropsychological testing. Additional studies using PET have suggested dysfunction of the cortical-thalamic-cerebellar-cortical circuit during working memory tasks (72, 73). The “cognitive dysmetria” resulting from this disruption may provide another candidate endophenotype.
Conclusions: Broader Uses for Endophenotypes
Endophenotypes may have additional uses in psychiatry, including uses in diagnosis, classification, and the development of animal models. The current classification schema in psychiatry were derived from observable clinical grounds to address the need for clinical description and communication (22). However, they are not based on measures of the underlying genetic or biological pathophysiology of the disorders. The most widely used systems currently in place must serve the needs of clinicians, psychiatric statisticians, administrators, and insurance companies, among other groups and agencies (128). As this system is designed for a wide range of users and because it pays little attention to the biological contributors to the disorders, it is not optimized for the design, implementation, and success of research studies (128). The lack of a biological basis for the classification of psychiatric disorders has led, in part, to a lack of success in studies of the neurobiology and genetics of psychiatric disorders. Endophenotype-based analysis would be useful for establishing a biological underpinning for diagnosis and classification; a net outcome would be improved understanding of the neurobiology and genetics of psychopathology.
Animal models are an active area of research in psychiatry. However, despite some progress (129, 130), there remains a great need for further development (130–132). Improved animal models will help in understanding the neurobiology of psychiatric disorders and will further the development of truly novel medications (133). Development of animal partial-models in psychiatry relies on identifying critical components of behavior (or other neurobiological traits) that are representative of more complex phenomena (134). Animals will never have guilty ruminations, suicidal thoughts, or rapid speech. Thus, animal models based on endophenotypes that represent evolutionarily selected and quantifiable traits may better lend themselves to investigation of psychiatric phenomena than models based on face-valid diagnostic phenotypes (28).
Given the hopefully successful consequences of studies adopting an endophenotype strategy, psychiatric diagnosis will continue to be important in research and clinical practice. Indeed, similar to the principle we describe here, optimally reduced or partitioned phenotypes may be useful in refining the diagnostic system. Measures that have already been used to deconstruct illnesses for genetic analysis include severity and course of illness (135), age at onset of illness (136, 137), amount of substance use in drug and alcohol disorders (138, 139), and response to specific treatments such as lithium (140, 141).
Gottesman and Shields (25) concluded their 1972 book on schizophrenia and genetics with the following remarks:
We are optimistically hopeful that the current mass of research on families of schizophrenics will discover an endophenotype, either biological or behavioral (psychometric pattern),which will not only discriminate schizophrenics from other psychotics, but will also be found in all the identical co-twins of schizophrenics whether concordant or discordant. All genetic theorizing will benefit from the development of such an indicator (p. 336).
Although these words are still pertinent after 30 years, there is ample reason to be optimistic about anticipated discoveries and refinements in the quest for endophenotypes.
From the Department of Psychiatry, University of Minnesota Medical School; and the Laboratory of Molecular Pathophysiology, Mood and Anxiety Disorders Program, NIMH, Bethesda, Md. Address reprint requests to Dr. Gottesman, Department of Psychiatry, University of Minnesota Medical School, 2450 Riverside Ave., Minneapolis, MN 55454; [email protected] (e-mail). Dr. Gould is supported by the NIMH intramural research program. The authors thank David A. Lewis, M.D., Husseini K. Manji, M.D., and Arturas Petronis, M.D., Ph.D., for their encouragement.
Figure 1. Rationale for an Endophenotype Approach to Genetic Analysis of Disorders With Complex Geneticsa
aThe number of genes involved in a phenotype is theorized to be directly related to both the complexity of the phenotype and the difficulty of genetic analysis (28–34).
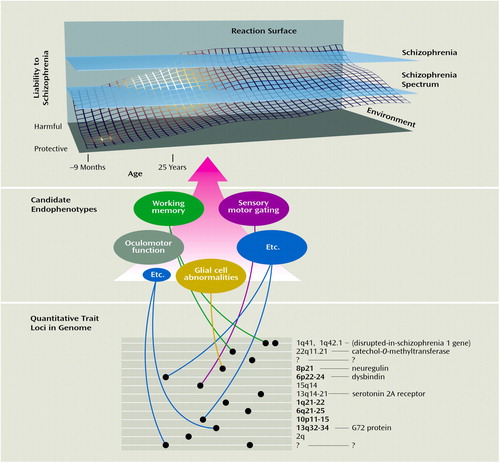
Figure 2. Gene Regions, Genes, and Putative Endophenotypes Implicated in a Biological Systems Approach to Schizophrenia Researcha
aThe reaction surface (36) suggests the dynamic developmental interplay among genetic, environmental, and epigenetic factors that produce cumulative liability to developing schizophrenia (9–11, 37). Gene regions where linkage findings are more consistent are in bold, while gene regions corresponding to candidate genes or endophenotypes are shown in normal lettering (16). Many of these endophenotypes are discussed in detailed reviews addressing overall strategies for schizophrenia discriminators (38), sensory motor gating (33, 39, 40), oculomotor function (33, 40–43), working memory (sometimes synonymous with information processing, executive function, attention) (31, 32, 44–46), and glial cell abnormalities (47). None of the sections of this figure can be definitive; many more gene loci, genes, and candidate endophenotypes exist and remain to be discovered (represented by question marks) (47, 48). Linkage and candidate gene studies have been the topic of recent reviews (15, 16, 49, 50). The figure is not to scale. (Copyright 2003, I.I. Gottesman. Used with permission.)
1. Sturtevant AH: A History of Genetics. New York, Cold Spring Harbor Laboratory Press, 1965 (reprinted 2001) (full text available online at http://www.esp.org/books/sturt/history)Google Scholar
2. Zerba KE, Ferrell RE, Sing CF: Complex adaptive systems and human health: the influence of common genotypes of the apolipoprotein E (ApoE) gene polymorphism and age on the relational order within a field of lipid metabolism traits. Hum Genet 2000; 107:466-475Crossref, Medline, Google Scholar
3. Sing CF, Zerba KE, Reilly SL: Traversing the biological complexity in the hierarchy between genome and CAD endpoints in the population at large. Clin Genet 1994; 46(1 special number):6-14Google Scholar
4. Province MA, Shannon WD, Rao DC: Classification methods for confronting heterogeneity. Adv Genet 2001; 42:273-286Crossref, Medline, Google Scholar
5. Gottesman II: Psychopathology through a life span-genetic prism. Am Psychol 2001; 56:867-878Crossref, Medline, Google Scholar
6. Merikangas KR, Swendsen JD: Genetic epidemiology of psychiatric disorders. Epidemiol Rev 1997; 19:144-155Crossref, Medline, Google Scholar
7. Petronis A, Gottesman, II, Crow TJ, DeLisi LE, Klar AJ, Macciardi F, McInnis MG, McMahon FJ, Paterson AD, Skuse D, Sutherland GR: Psychiatric epigenetics: a new focus for the new century. Mol Psychiatry 2000; 5:342-346Crossref, Medline, Google Scholar
8. Rakyan VK, Preis J, Morgan HD, Whitelaw E: The marks, mechanisms and memory of epigenetic states in mammals. Biochem J 2001; 356(part 1):1-10Google Scholar
9. McGuffin P, Owen MJ, Gottesman II: Psychiatric Genetics and Genomics. Oxford, UK, Oxford University Press, 2002Google Scholar
10. Lewis DA, Levitt P: Schizophrenia as a disorder of neurodevelopment. Annu Rev Neurosci 2002; 25:409-432Crossref, Medline, Google Scholar
11. Petronis A: Human morbid genetics revisited: relevance of epigenetics. Trends Genet 2001; 17:142-146Crossref, Medline, Google Scholar
12. Glazier AM, Nadeau JH, Aitman TJ: Finding genes that underlie complex traits. Science 2002; 298:2345-2349Crossref, Medline, Google Scholar
13. Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, et al: The sequence of the human genome. Science 2001; 291:1304-1305; correction, 2001; 292:1838Google Scholar
14. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, et al: Initial sequencing and analysis of the human genome. Nature 2001; 409:860-921Crossref, Medline, Google Scholar
15. Cowan WM, Kopnisky KL, Hyman SE: The human genome project and its impact on psychiatry. Annu Rev Neurosci 2002; 25:1-50Crossref, Medline, Google Scholar
16. Sklar P: Linkage analysis in psychiatric disorders: the emerging picture. Annu Rev Genomics Hum Genet 2002; 3:371-413Crossref, Medline, Google Scholar
17. Gottesman II, Moldin SO: Schizophrenia genetics at the millennium: cautious optimism. Clin Genet 1997; 52:404-407Crossref, Medline, Google Scholar
18. Cloninger CR: The discovery of susceptibility genes for mental disorders. Proc Natl Acad Sci USA 2002; 99:13365-13367Crossref, Medline, Google Scholar
19. Andreasen NC: Understanding the causes of schizophrenia (letter). N Engl J Med 1999; 340:645-647Crossref, Medline, Google Scholar
20. Lewis DA: In pursuit of the pathogenesis and pathophysiology of schizophrenia: where do we stand? (editorial). Am J Psychiatry 2002; 159:1467-1469Link, Google Scholar
21. Chakravarti A, Little P: Nature, nurture and human disease. Nature 2003; 421:412-414Crossref, Medline, Google Scholar
22. Andreasen NC: Schizophrenia: the fundamental questions. Brain Res Brain Res Rev 2000; 31:106-112Crossref, Medline, Google Scholar
23. Kandel ER: A new intellectual framework for psychiatry. Am J Psychiatry 1998; 155:457-469Link, Google Scholar
24. Gottesman, II, Shields J: A polygenic theory of schizophrenia. Proc Natl Acad Sci USA 1967; 58:199-205Crossref, Medline, Google Scholar
25. Gottesman II, Shields J: Schizophrenia and Genetics: A Twin Study Vantage Point. New York, Academic Press, 1972Google Scholar
26. Gottesman II, Shields J: Genetic theorizing and schizophrenia. Br J Psychiatry 1973; 122:15-30Crossref, Medline, Google Scholar
27. John B, Lewis KR: Chromosome variability and geographical distribution in insects: chromosome rather than gene variation provide the key to differences among populations. Science 1966; 152:711-721Crossref, Medline, Google Scholar
28. Lenox RH, Gould TD, Manji HK: Endophenotypes in bipolar disorder. Am J Med Genet 2002; 114:391-406Crossref, Medline, Google Scholar
29. Leboyer M, Bellivier F, Nosten-Bertrand M, Jouvent R, Pauls D, Mallet J: Psychiatric genetics: search for phenotypes. Trends Neurosci 1998; 21:102-105Crossref, Medline, Google Scholar
30. Callicott JH, Weinberger DR: Brain imaging as an approach to phenotype characterization for genetic studies of schizophrenia. Methods Mol Med 2003; 77:227-247Medline, Google Scholar
31. Weinberger DR, Egan MF, Bertolino A, Callicott JH, Mattay VS, Lipska BK, Berman KF, Goldberg TE: Prefrontal neurons and the genetics of schizophrenia. Biol Psychiatry 2001; 50:825-844Crossref, Medline, Google Scholar
32. Egan MF, Goldberg TE: Intermediate cognitive phenotypes associated with schizophrenia. Methods Mol Med 2003; 77:163-197Medline, Google Scholar
33. Braff DL, Freedman R: Endophenotypes in studies of the genetics of schizophrenia, in Neuropsychopharmacology: The Fifth Generation of Progress. Edited by Davis KL, Charney DS, Coyle JT, Nemeroff C. Philadelphia, Lippincott Williams & Wilkins, 2002, pp 703-716Google Scholar
34. Leboyer M: Searching for alternative phenotypes in psychiatric genetics. Methods Mol Med 2003; 77:145-161Medline, Google Scholar
35. MacCorquodale K, Meehl PE: On a distinction between hypothetical constructs and intervening variables. Psychol Rev 1948; 55:95-107Crossref, Medline, Google Scholar
36. Turkheimer E, Goldsmith HH, Gottesman II: Commentary: some conceptual deficiencies in “developmental” behavior genetics. Hum Dev 1995; 38:142-153Crossref, Google Scholar
37. Faraone SV, Tsuang D, Tsuang MT: Genetics of Mental Disorders: A Guide for Students, Clinicians, and Researchers. New York, Guilford, 1999Google Scholar
38. Heinrichs RW: In Search of Madness: Schizophrenia and Neuroscience. New York, Oxford University Press, 2001Google Scholar
39. Braff DL, Geyer MA, Swerdlow NR: Human studies of prepulse inhibition of startle: normal subjects, patient groups, and pharmacological studies. Psychopharmacology (Berl) 2001; 156:234-258Crossref, Medline, Google Scholar
40. Freedman R: Electrophysiological phenotypes. Methods Mol Med 2003; 77:215-225Medline, Google Scholar
41. Holzman PS: Less is truly more: psychopathology research in the 21st century, in Principles of Psychopathology: Essays in Honor of Brendan A. Maher. Edited by Lenzenweger MF, Hooley JM. Washington, American Psychological Association, 2003, pp 175-194Google Scholar
42. Calkins ME, Iacono WG: Eye movement dysfunction in schizophrenia: a heritable characteristic for enhancing phenotype definition. Am J Med Genet 2000; 97:72-76Crossref, Medline, Google Scholar
43. Lee KH, Williams LM: Eye movement dysfunction as a biological marker of risk for schizophrenia. Aust N Z J Psychiatry 2000; 34(Suppl):S91-S100Google Scholar
44. Goldberg TE, Green MF: Neurocognitive functioning in patients with schizophrenia: an overview, in Neuropsychopharmacology: The Fifth Generation of Progress. Edited by Davis KL, Charney DS, Coyle JT, Nemeroff C. Philadelphia, Lippincott Williams & Wilkins, 2002, pp 657-669Google Scholar
45. Goldman-Rakic PS: The physiological approach: functional architecture of working memory and disordered cognition in schizophrenia. Biol Psychiatry 1999; 46:650-661Crossref, Medline, Google Scholar
46. Erlenmeyer-Kimling L, Rock D, Roberts SA, Janal M, Kestenbaum C, Cornblatt B, Adamo UH, Gottesman II: Attention, memory, and motor skills as childhood predictors of schizophrenia-related psychoses: the New York High-Risk Project. Am J Psychiatry 2000; 157:1416-1422Link, Google Scholar
47. Moises HW, Zoega T, Gottesman II: The glial growth factors deficiency and synaptic destabilization hypothesis of schizophrenia. BMC Psychiatry 2002; 2:8, http://www.biomedcentral. com/1471-244X/2/8/Crossref, Medline, Google Scholar
48. Wise LH, Lanchbury JS, Lewis CM: Meta-analysis of genome searches. Ann Hum Genet 1999; 63(part 3):263-272Google Scholar
49. Owen MJ, O’Donovan MC, Gottesman II: Schizophrenia, in Psychiatric Genetics and Genomics. Edited by McGuffin P, Owens MJ, Gottesman II. Oxford, UK, Oxford University Press, 2002, pp 247-266Google Scholar
50. Pulver AE, Pearlson G, McGrath J, Lasseter VK, Swarts K, Papadimitriou G: Schizophrenia, in The Genetic Basis of Common Diseases. Edited by King RA, Rotter JI, Motulsky AG. New York, Oxford University Press, 2002Google Scholar
51. Trumbetta SL, Gottesman II: Endophenotypes for marital status in the NAS-NRC Twin Registry, in Genetic Influences on Human Fertility and Sexuality. Edited by Rodgers JL, Rowe DC, Miller WB. Boston, Kluwer Academic, 2000, pp 253-269Google Scholar
52. Skuse DH: Endophenotypes and child psychiatry. Br J Psychiatry 2001; 178:395-396Crossref, Medline, Google Scholar
53. Gasperoni TL, Ekelund J, Huttunen M, Palmer CG, Tuulio-Henriksson A, Lonnqvist J, Kaprio J, Peltonen L, Cannon TD: Genetic linkage and association between chromosome 1q and working memory function in schizophrenia. Am J Med Genet 2003; 116(1 suppl):8-16Google Scholar
54. Castellanos FX, Tannock R: Neuroscience of attention-deficit/hyperactivity disorder: the search for endophenotypes. Nat Rev Neurosci 2002; 3:617-628Crossref, Medline, Google Scholar
55. Ahearn EP, Speer MC, Chen YT, Steffens DC, Cassidy F, Van Meter S, Provenzale JM, Weisler RH, Krishnan KR: Investigation of Notch3 as a candidate gene for bipolar disorder using brain hyperintensities as an endophenotype. Am J Med Genet 2002; 114:652-658Crossref, Medline, Google Scholar
56. Cadenhead KS, Light GA, Geyer MA, McDowell JE, Braff DL: Neurobiological measures of schizotypal personality disorder: defining an inhibitory endophenotype? Am J Psychiatry 2002; 159:869-871Link, Google Scholar
57. Cornblatt BA, Malhotra AK: Impaired attention as an endophenotype for molecular genetic studies of schizophrenia. Am J Med Genet 2001; 105:11-15Crossref, Medline, Google Scholar
58. Merikangas KR, Chakravarti A, Moldin SO, Araj H, Blangero JC, Burmeister M, Crabbe J Jr, Depaulo JR Jr, Foulks E, Freimer NB, Koretz DS, Lichtenstein W, Mignot E, Reiss AL, Risch NJ, Takahashi JS: Future of genetics of mood disorders research. Biol Psychiatry 2002; 52:457-477Crossref, Medline, Google Scholar
59. Seibyl JP, Scanley E, Krystal JH, Innis RB: Neuroimaging methodologies, in Neurobiology of Mental Illness. Edited by Charney DS, Nester EJ, Bunney BS. New York, Oxford University Press, 1999, pp 170-189Google Scholar
60. Diwadkar VA, Keshavan MS: Newer techniques in magnetic resonance imaging and their potential for neuropsychiatric research. J Psychosom Res 2002; 53:677-685Crossref, Medline, Google Scholar
61. Martinez D, Broft A, Laruelle M: Imaging neurochemical endophenotypes: promises and pitfalls. Pharmacogenomics 2001; 2:223-237Crossref, Medline, Google Scholar
62. Gershon ES, Goldin LR: Clinical methods in psychiatric genetics, I: robustness of genetic marker investigative strategies. Acta Psychiatr Scand 1986; 74:113-118Crossref, Medline, Google Scholar
63. Keating MT, Sanguinetti MC: Molecular and cellular mechanisms of cardiac arrhythmias. Cell 2001; 104:569-580Crossref, Medline, Google Scholar
64. Keating M, Atkinson D, Dunn C, Timothy K, Vincent GM, Leppert M: Linkage of a cardiac arrhythmia, the long QT syndrome, and the Harvey ras-1 gene. Science 1991; 252:704-706Crossref, Medline, Google Scholar
65. Vincent GM, Timothy KW, Leppert M, Keating M: The spectrum of symptoms and QT intervals in carriers of the gene for the long-QT syndrome. N Engl J Med 1992; 327:846-852Crossref, Medline, Google Scholar
66. Casimiro MC, Knollmann BC, Ebert SN, Vary JC Jr, Greene AE, Franz MR, Grinberg A, Huang SP, Pfeifer K: Targeted disruption of the Kcnq1 gene produces a mouse model of Jervell and Lange-Nielsen syndrome. Proc Natl Acad Sci USA 2001; 98:2526-2531Crossref, Medline, Google Scholar
67. Lalouel JM, Le Mignon L, Simon M, Fauchet R, Bourel M, Rao DC, Morton NE: Genetic analysis of idiopathic hemochromatosis using both qualitative (disease status) and quantitative (serum iron) information. Am J Hum Genet 1985; 37:700-718Medline, Google Scholar
68. Greenberg DA, Delgado-Escueta AV, Widelitz H, Sparkes RS, Treiman L, Maldonado HM, Park MS, Terasaki PI: Juvenile myoclonic epilepsy (JME) may be linked to the BF and HLA loci on human chromosome 6. Am J Med Genet 1988; 31:185-192Crossref, Medline, Google Scholar
69. Leppert M, Burt R, Hughes JP, Samowitz W, Nakamura Y, Woodward S, Gardner E, Lalouel JM, White R: Genetic analysis of an inherited predisposition to colon cancer in a family with a variable number of adenomatous polyps. N Engl J Med 1990; 322:904-908Crossref, Medline, Google Scholar
70. Gottesman II, Erlenmeyer-Kimling L: Family and twin strategies as a head start in defining prodromes and endophenotypes for hypothetical early-interventions in schizophrenia. Schizophr Res 2001; 51:93-102Crossref, Medline, Google Scholar
71. Lenzenweger MF: Schizophrenia: refining the phenotype, resolving endophenotypes. Behav Res Ther 1999; 37:281-295Crossref, Medline, Google Scholar
72. Andreasen NC, O"Leary DS, Cizadlo T, Arndt S, Rezai K, Ponto LL, Watkins GL, Hichwa RD: Schizophrenia and cognitive dysmetria: a positron-emission tomography study of dysfunctional prefrontal-thalamic-cerebellar circuitry. Proc Natl Acad Sci U S A 1996; 93:9985-9990Crossref, Medline, Google Scholar
73. Andreasen NC, Nopoulos P, O"Leary DS, Miller DD, Wassink T, Flaum M: Defining the phenotype of schizophrenia: cognitive dysmetria and its neural mechanisms. Biol Psychiatry 1999; 46:908-920Crossref, Medline, Google Scholar
74. Berman RM, Narasimhan M, Miller HL, Anand A, Cappiello A, Oren DA, Heninger GR, Charney DS: Transient depressive relapse induced by catecholamine depletion: potential phenotypic vulnerability marker? Arch Gen Psychiatry 1999; 56:395-403Crossref, Medline, Google Scholar
75. Niculescu AB III, Akiskal HS: Proposed endophenotypes of dysthymia: evolutionary, clinical and pharmacogenomic considerations. Mol Psychiatry 2001; 6:363-366Crossref, Medline, Google Scholar
76. Neugroschl J, Davis KL: Biological markers in Alzheimer disease. Am J Geriatr Psychiatry 2002; 10:660-677Crossref, Medline, Google Scholar
77. Kurz A, Riemenschneider M, Drzezga A, Lautenschlager N: The role of biological markers in the early and differential diagnosis of Alzheimer’s disease. J Neural Transm Suppl 2002:127-133Google Scholar
78. Gould TD, Bastain TM, Israel ME, Hommer DW, Castellanos FX: Altered performance on an ocular fixation task in attention-deficit/hyperactivity disorder. Biol Psychiatry 2001; 50:633-635Crossref, Medline, Google Scholar
79. Seidman LJ, Biederman J, Monuteaux MC, Weber W, Faraone SV: Neuropsychological functioning in nonreferred siblings of children with attention deficit/hyperactivity disorder. J Abnorm Psychol 2000; 109:252-265Crossref, Medline, Google Scholar
80. New AS, Siever LJ: Biochemical endophenotypes in personality disorders. Methods Mol Med 2003; 77:199-213Medline, Google Scholar
81. Braff DL, Geyer MA: Sensorimotor gating and schizophrenia: human and animal model studies. Arch Gen Psychiatry 1990; 47:181-188Crossref, Medline, Google Scholar
82. Grillon C, Courchesne E, Ameli R, Geyer MA, Braff DL: Increased distractibility in schizophrenic patients: electrophysiologic and behavioral evidence. Arch Gen Psychiatry 1990; 47:171-179Crossref, Medline, Google Scholar
83. McGhie A, Chapman J: Disorders of attention and perception in early schizophrenia. Br J Med Psychol 1961; 34:103-116Crossref, Medline, Google Scholar
84. Braff DL: Psychophysiological and information-processing approaches to schizophrenia, in Neurobiology of Mental Illness. Edited by Charney DS, Nester EJ, Bunney BS. New York, Oxford University Press, 1999, pp 258-271Google Scholar
85. Braff DL, Grillon C, Geyer MA: Gating and habituation of the startle reflex in schizophrenic patients. Arch Gen Psychiatry 1992; 49:206-215Crossref, Medline, Google Scholar
86. Geyer MA, McIlwain KL, Paylor R: Mouse genetic models for prepulse inhibition: an early review. Mol Psychiatry 2002; 7:1039-1053Crossref, Medline, Google Scholar
87. Cadenhead KS, Swerdlow NR, Shafer KM, Diaz M, Braff DL: Modulation of the startle response and startle laterality in relatives of schizophrenic patients and in subjects with schizotypal personality disorder: evidence of inhibitory deficits. Am J Psychiatry 2000; 157:1660-1668; correction, 157:1904Link, Google Scholar
88. Ellenbroek BA, Cools AR: Early maternal deprivation and prepulse inhibition: the role of the postdeprivation environment. Pharmacol Biochem Behav 2002; 73:177-184Crossref, Medline, Google Scholar
89. Weiss IC, Feldon J: Environmental animal models for sensorimotor gating deficiencies in schizophrenia: a review. Psychopharmacology (Berl) 2001; 156:305-326Crossref, Medline, Google Scholar
90. Swerdlow NR, Benbow CH, Zisook S, Geyer MA, Braff DL: A preliminary assessment of sensorimotor gating in patients with obsessive compulsive disorder. Biol Psychiatry 1993; 33:298-301Crossref, Medline, Google Scholar
91. Swerdlow NR, Paulsen J, Braff DL, Butters N, Geyer MA, Swenson MR: Impaired prepulse inhibition of acoustic and tactile startle response in patients with Huntington’s disease. J Neurol Neurosurg Psychiatry 1995; 58:192-200Crossref, Medline, Google Scholar
92. Adler LE, Pachtman E, Franks RD, Pecevich M, Waldo MC, Freedman R: Neurophysiological evidence for a defect in neuronal mechanisms involved in sensory gating in schizophrenia. Biol Psychiatry 1982; 17:639-654Medline, Google Scholar
93. Freedman R, Adler LE, Waldo MC, Pachtman E, Franks RD: Neurophysiological evidence for a defect in inhibitory pathways in schizophrenia: comparison of medicated and drug-free patients. Biol Psychiatry 1983; 18:537-551Medline, Google Scholar
94. Freedman R, Adler LE, Gerhardt GA, Waldo M, Baker N, Rose GM, Drebing C, Nagamoto H, Bickford-Wimer P, Franks R: Neurobiological studies of sensory gating in schizophrenia. Schizophr Bull 1987; 13:669-678Crossref, Medline, Google Scholar
95. Siegel C, Waldo M, Mizner G, Adler LE, Freedman R: Deficits in sensory gating in schizophrenic patients and their relatives: evidence obtained with auditory evoked responses. Arch Gen Psychiatry 1984; 41:607-612Crossref, Medline, Google Scholar
96. Myles-Worsley M: P50 sensory gating in multiplex schizophrenia families from a Pacific Island isolate. Am J Psychiatry 2002; 159:2007-2012Link, Google Scholar
97. Clementz BA, Geyer MA, Braff DL: Poor P50 suppression among schizophrenia patients and their first-degree biological relatives. Am J Psychiatry 1998; 155:1691-1694Link, Google Scholar
98. Waldo MC, Carey G, Myles-Worsley M, Cawthra E, Adler LE, Nagamoto HT, Wender P, Byerley W, Plaetke R, Freedman R: Codistribution of a sensory gating deficit and schizophrenia in multi-affected families. Psychiatry Res 1991; 39:257-268Crossref, Medline, Google Scholar
99. Waldo M, Myles-Worsley M, Madison A, Byerley W, Freedman R: Sensory gating deficits in parents of schizophrenics. Am J Med Genet 1995; 60:506-511Crossref, Medline, Google Scholar
100. Myles-Worsley M, Coon H, Byerley W, Waldo M, Young D, Freedman R: Developmental and genetic influences on the P50 sensory gating phenotype. Biol Psychiatry 1996; 39:289-295Crossref, Medline, Google Scholar
101. Young DA, Waldo M, Rutledge JH III, Freedman R: Heritability of inhibitory gating of the P50 auditory-evoked potential in monozygotic and dizygotic twins. Neuropsychobiology 1996; 33:113-117Crossref, Medline, Google Scholar
102. Freedman R, Coon H, Myles-Worsley M, Orr-Urtreger A, Olincy A, Davis A, Polymeropoulos M, Holik J, Hopkins J, Hoff M, Rosenthal J, Waldo MC, Reimherr F, Wender P, Yaw J, Young DA, Breese CR, Adams C, Patterson D, Adler LE, Kruglyak L, Leonard S, Byerley W: Linkage of a neurophysiological deficit in schizophrenia to a chromosome 15 locus. Proc Natl Acad Sci USA 1997; 94:587-592Crossref, Medline, Google Scholar
103. Freedman R, Leonard S, Gault JM, Hopkins J, Cloninger CR, Kaufmann CA, Tsuang MT, Faraone SV, Malaspina D, Svrakic DM, Sanders A, Gejman P: Linkage disequilibrium for schizophrenia at the chromosome 15q13-14 locus of the α7-nicotinic acetylcholine receptor subunit gene (CHRNA7). Am J Med Genet 2001; 105:20-22Crossref, Medline, Google Scholar
104. Leonard S, Gault J, Hopkins J, Logel J, Vianzon R, Short M, Drebing C, Berger R, Venn D, Sirota P, Zerbe G, Olincy A, Ross RG, Adler LE, Freedman R: Association of promoter variants in the α7 nicotinic acetylcholine receptor subunit gene with an inhibitory deficit found in schizophrenia. Arch Gen Psychiatry 2002; 59:1085-1096Crossref, Medline, Google Scholar
105. Diefendorf AR, Dodge R: An experimental study of the ocular reactions of the insane from photographic records. Brain 1908; 31:451-489Crossref, Google Scholar
106. Holzman PS, Proctor LR, Hughes DW: Eye-tracking patterns in schizophrenia. Science 1973; 181:179-181Crossref, Medline, Google Scholar
107. Holzman PS, Proctor LR, Levy DL, Yasillo NJ, Meltzer HY, Hurt SW: Eye-tracking dysfunctions in schizophrenic patients and their relatives. Arch Gen Psychiatry 1974; 31:143-151Crossref, Medline, Google Scholar
108. Munoz DP: Commentary: saccadic eye movements: overview of neural circuitry. Prog Brain Res 2002; 140:89-96Crossref, Medline, Google Scholar
109. Arolt V, Lencer R, Nolte A, Muller-Myhsok B, Purmann S, Schurmann M, Leutelt J, Pinnow M, Schwinger E: Eye tracking dysfunction is a putative phenotypic susceptibility marker of schizophrenia and maps to a locus on chromosome 6p in families with multiple occurrence of the disease. Am J Med Genet 1996; 67(6):564-579Google Scholar
110. O"Driscoll GA, Benkelfat C, Florencio PS, Wolff AL, Joober R, Lal S, Evans AC: Neural correlates of eye tracking deficits in first-degree relatives of schizophrenic patients: a positron emission tomography study. Arch Gen Psychiatry 1999; 56:1127-1134Crossref, Medline, Google Scholar
111. Park S, Lee J: Spatial working memory function in schizophrenia, in Principles of Psychopathology: Essays in Honor of Brendan A. Maher. Edited by Lenzenweger MF, Hooley JM. Washington, American Psychological Association, 2003, pp 83-106Google Scholar
112. Snitz BE, Curtis CE, Zald DH, Katsanis J, Iacono WG: Neuropsychological and oculomotor correlates of spatial working memory performance in schizophrenia patients and controls. Schizophr Res 1999; 38(1):37-50Google Scholar
113. Levy R, Goldman-Rakic PS: Segregation of working memory functions within the dorsolateral prefrontal cortex. Exp Brain Res 2000; 133:23-32Crossref, Medline, Google Scholar
114. Harrison PJ: The neuropathology of schizophrenia: a critical review of the data and their interpretation. Brain 1999; 122(part 4):593-624Google Scholar
115. Park S, Holzman PS, Goldman-Rakic PS: Spatial working memory deficits in the relatives of schizophrenic patients. Arch Gen Psychiatry 1995; 52:821-828Crossref, Medline, Google Scholar
116. Conklin HM, Curtis CE, Katsanis J, Iacono WG: Verbal working memory impairment in schizophrenia patients and their first-degree relatives: evidence from the Digit Span Task. Am J Psychiatry 2000; 157:275-277Link, Google Scholar
117. Cannon TD, Huttunen MO, Lonnqvist J, Tuulio-Henriksson A, Pirkola T, Glahn D, Finkelstein J, Hietanen M, Kaprio J, Koskenvuo M: The inheritance of neuropsychological dysfunction in twins discordant for schizophrenia. Am J Hum Genet 2000; 67:369-382Crossref, Medline, Google Scholar
118. Goldberg TE, Kelsoe JR, Weinberger DR, Pliskin NH, Kirwin PD, Berman KF: Performance of schizophrenic patients on putative neuropsychological tests of frontal lobe function. Int J Neurosci 1988; 42:51-58Crossref, Medline, Google Scholar
119. Hovatta I, Varilo T, Suvisaari J, Terwilliger JD, Ollikainen V, Arajarvi R, Juvonen H, Kokko-Sahin ML, Vaisanen L, Mannila H, Lonnqvist J, Peltonen L: A genomewide screen for schizophrenia genes in an isolated Finnish subpopulation, suggesting multiple susceptibility loci. Am J Hum Genet 1999; 65:1114-1124Crossref, Medline, Google Scholar
120. Ekelund J, Lichtermann D, Hovatta I, Ellonen P, Suvisaari J, Terwilliger JD, Juvonen H, Varilo T, Arajarvi R, Kokko-Sahin ML, Lonnqvist J, Peltonen L: Genome-wide scan for schizophrenia in the Finnish population: evidence for a locus on chromosome 7q22. Hum Mol Genet 2000; 9:1049-1057Crossref, Medline, Google Scholar
121. St Clair D, Blackwood D, Muir W, Carothers A, Walker M, Spowart G, Gosden C, Evans HJ: Association within a family of a balanced autosomal translocation with major mental illness. Lancet 1990; 336:13-16Crossref, Medline, Google Scholar
122. Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, Clair DM, Muir WJ, Blackwood DH, Porteous DJ: Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet 2000; 9:1415-1423Crossref, Medline, Google Scholar
123. Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE, Goldman D, Weinberger DR: Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci USA 2001; 98:6917-6922Crossref, Medline, Google Scholar
124. Manoach DS, Gollub RL, Benson ES, Searl MM, Goff DC, Halpern E, Saper CB, Rauch SL: Schizophrenic subjects show aberrant fMRI activation of dorsolateral prefrontal cortex and basal ganglia during working memory performance. Biol Psychiatry 2000; 48:99-109Crossref, Medline, Google Scholar
125. Manoach DS, Press DZ, Thangaraj V, Searl MM, Goff DC, Halpern E, Saper CB, Warach S: Schizophrenic subjects activate dorsolateral prefrontal cortex during a working memory task, as measured by fMRI. Biol Psychiatry 1999; 45:1128-1137Crossref, Medline, Google Scholar
126. Callicott JH, Bertolino A, Mattay VS, Langheim FJ, Duyn J, Coppola R, Goldberg TE, Weinberger DR: Physiological dysfunction of the dorsolateral prefrontal cortex in schizophrenia revisited. Cereb Cortex 2000; 10:1078-1092Crossref, Medline, Google Scholar
127. Callicott JH, Egan MF, Mattay VS, Bertolino A, Bone AD, Verchinksi B, Weinberger DR: Abnormal fMRI response of the dorsolateral prefrontal cortex in cognitively intact siblings of patients with schizophrenia. Am J Psychiatry 2003; 160:709-719Link, Google Scholar
128. Duffy A, Grof P: Psychiatric diagnoses in the context of genetic studies of bipolar disorder. Bipolar Disord 2001; 3:270-275Crossref, Medline, Google Scholar
129. Crawley JN: What’s Wrong With My Mouse? Behavioral Phenotyping of Transgenic and Knockout Mice. New York, Wiley-Liss, 2000Google Scholar
130. Einat H, Belmaker RH, Manji HK: New approaches to modeling bipolar disorder. Psychopharmacol Bull (in press)Google Scholar
131. Nestler EJ, Gould E, Manji H, Buncan M, Duman RS, Greshenfeld HK, Hen R, Koester S, Lederhendler I, Meaney M, Robbins T, Winsky L, Zalcman S: Preclinical models: status of basic research in depression. Biol Psychiatry 2002; 52:503-528Crossref, Medline, Google Scholar
132. Einat H, Kofman O, Belmaker RH: Animal models of bipolar disorder: from a single episode to progressive cycling models, in Contemporary Issues in Modeling Psychopharmacology. Edited by Weiner I. Boston, Kluwer Academic, 2000, pp 165-180Google Scholar
133. Gould TD, Gray NA, Manji HK: The cellular neurobiology of severe mood and anxiety disorders: implications for the development of novel therapeutics, in Molecular Neurobiology for the Clinician. Edited by Charney DS. Washington, American Psychiatric Publishing (in press)Google Scholar
134. Crawley JN: Behavioral phenotyping of transgenic and knockout mice: experimental design and evaluation of general health, sensory functions, motor abilities, and specific behavioral tests. Brain Res 1999; 835:18-26Crossref, Medline, Google Scholar
135. Geller B, Cook EH Jr: Ultradian rapid cycling in prepubertal and early adolescent bipolarity is not in transmission disequilibrium with val/met COMT alleles. Biol Psychiatry 2000; 47:605-609Crossref, Medline, Google Scholar
136. Cardno AG, Holmans PA, Rees MI, Jones LA, McCarthy GM, Hamshere ML, Williams NM, Norton N, Williams HJ, Fenton I, Murphy KC, Sanders RD, Gray MY, O’Donovan MC, McGuffin P, Owen MJ: A genomewide linkage study of age at onset in schizophrenia. Am J Med Genet 2001; 105:439-445Crossref, Medline, Google Scholar
137. Zubenko GS, Hughes HB, Stiffler JS, Zubenko WN, Kaplan BB: Genome survey for susceptibility loci for recurrent, early-onset major depression: results at 10cM resolution. Am J Med Genet 2002; 114:413-422Crossref, Medline, Google Scholar
138. Wall TL, Carr LG, Ehlers CL: Protective association of genetic variation in alcohol dehydrogenase with alcohol dependence in Native American Mission Indians. Am J Psychiatry 2003; 160:41-46Link, Google Scholar
139. Saccone NL, Kwon JM, Corbett J, Goate A, Rochberg N, Edenberg HJ, Foroud T, Li TK, Begleiter H, Reich T, Rice JP: A genome screen of maximum number of drinks as an alcoholism phenotype. Am J Med Genet 2000; 96:632-637Crossref, Medline, Google Scholar
140. Mansour HA, Alda M, Nimgaonkar VL: Pharmacogenetics of bipolar disorder. Curr Psychiatry Rep 2002; 4:117-123Crossref, Medline, Google Scholar
141. Detera-Wadleigh SD: Lithium-related genetics of bipolar disorder. Ann Med 2001; 33:272-285Crossref, Medline, Google Scholar



