Genome-Wide Methylation Changes in the Brains of Suicide Completers
Abstract
Objective
Gene expression changes have been reported in the brains of suicide completers. More recently, differences in promoter DNA methylation between suicide completers and comparison subjects in specific genes have been associated with these changes in gene expression patterns, implicating DNA methylation alterations as a plausible component of the pathophysiology of suicide. The authors used a genome-wide approach to investigate the extent of DNA methylation alterations in the brains of suicide completers.
Method
Promoter DNA methylation was profiled using methylated DNA immunoprecipitation (MeDIP) followed by microarray hybridization in hippocampal tissue from 62 men (46 suicide completers and 16 comparison subjects). The correlation between promoter methylation and expression was investigated by comparing the MeDIP data with gene expression profiles generated through mRNA microarray. Methylation differences between groups were validated on neuronal and nonneuronal DNA fractions isolated by fluorescence-assisted cell sorting.
Results
The authors identified 366 promoters that were differentially methylated in suicide completers relative to comparison subjects (273 hypermethylated and 93 hypomethylated). Overall, promoter methylation differences were inversely correlated with gene expression differences. Functional annotation analyses revealed an enrichment of differential methylation in the promoters of genes involved, among other functions, in cognitive processes. Validation was performed on the top genes from this category, and these differences were found to occur mainly in the neuronal cell fraction.
Conclusions
These results suggest broad reprogramming of promoter DNA methylation patterns in the hippocampus of suicide completers. This may help explain gene expression alterations associated with suicide and possibly behavioral changes increasing suicide risk.
The World Health Organization has estimated that the burden of suicide is 20 million life-years and that in 2020 suicide could be responsible for 2.4% of the total burden of disease (1). Despite its major impact on society and the attention it increasingly receives, our understanding of suicide remains poor.
Suicide results from interactions between biological, developmental, and social factors, and studies suggest that individuals who die by suicide have a certain predisposition (2). Although suicide is strongly associated with depressive psychopathology, previous studies have clearly indicated that predisposition to suicide, while partially overlapping with vulnerability to depression, is distinct (3). Because suicide risk factors are likely to induce risk by acting on brain processes, delineating mechanisms by which these risk factors affect brain function is of high interest. Recent evidence suggests that alterations in DNA methylation may play a role in the neurobiological processes leading up to suicide.
DNA methylation is a covalent modification of the DNA molecule that plays a role in regulating genome function in conjunction with other mechanisms, such as histone modifications (4, 5). Although DNA methylation is part of the chemistry of the DNA molecule, it is dynamic and responsive to external cues and signals. As such, the suggestion has been made that it serves as a “genome adaptation” mechanism (6).
A growing body of evidence demonstrates the presence of epigenetic alterations in the brains of suicide completers. Relative to comparison populations, suicide completers show lower expression of the astrocytic variant of the tyrosine kinase receptor B (TrkB-T1) gene in association with promoter hypermethylation in the prefrontal cortex (7), site-specific hypermethylation in the promoter of the brain-derived neurotrophic factor variant IV (BDNF-IV) associated with decreased expression in the Wernicke area (8), and decreased expression of the GABA-ergic receptor alpha subunit (GABAα) gene with hypermethylation in its promoter in the prefrontal cortex (9). Interestingly, these associations were independent of the possible effect of psychopathology. More recently, hypermethylation in the promoter of the glucocorticoid receptor gene was associated with decreased expression of glucocorticoid receptor in the hippocampus of abused suicide completers (10, 11). These findings are supported by studies in animal models of maternal and stress-induced depressive-like behaviors showing DNA methylation alterations in animal brains (12).
Collectively, these data suggest that DNA-specific methylation patterns are associated with suicide, independently from psychopathology. To date, this hypothesis has been primarily investigated by candidate-gene studies. Our understanding of gene interactions, however, suggests that genes act in complex networks, and it therefore stands to reason that DNA methylation involved in complex pathophysiology leading to suicide is not limited to a few candidate genes. We report results from a comprehensive genome-wide screening of promoter DNA methylation modifications found in the hippocampus of suicide completers compared with nonpsychiatric sudden-death comparison subjects. The results support the hypothesis that promoter DNA methylation patterns are altered in a coordinated way, affecting gene expression throughout the genome.
Method
Full details of the study methods are provided in the data supplement that accompanies the online edition of this article.
Samples
The project was approved by the Research Ethics Board at the Douglas Mental Health University Institute, Verdun, Quebec. Brain tissues were obtained from the Quebec Suicide Brain Bank, Douglas Mental Health University Institute. Brain samples (dentate gyrus) from the left hemisphere were obtained from 62 subjects (46 suicide completers and 16 nonpsychiatric sudden-death comparison subjects; see Table S1 in the online data supplement). Samples were carefully dissected at 4°C after having been flash-frozen in isopentene at –80°C. Dissection of the hippocampus was performed by expert brain bank staff using reference neuroanatomical maps (13). All samples were from Caucasian males of French Canadian descent, a population with a well-identified founder effect (14), and were group-matched for age, pH, and postmortem interval. Age, postmortem interval, presence of adversity, and comorbidity in the suicide group were controlled for by regression analysis. (See the online data supplement, including Table S1, for a complete description of sample selection and demographic characteristics.)
Methylated DNA Immunoprecipitation, Labeling, and Hybridization
Methylated DNA was extracted from each sample following methylated DNA immunoprecipitation (meDIP) as previously described (15) using 5′ methylcystosine antibody. Every sample was hybridized on a separate microarray. Microarrays were scanned using an Agilent High-Resolution C Scanner (Santa Clara, Calif.). Data were extracted using Agilent’s Feature Extraction software. (See the online data supplement for complete descriptions of meDIP, labeling, and hybridization methods.)
DNA Methylation Microarrays
DNA was hybridized on custom-designed (eArray, July 2009) 400K promoter tiling microarrays (Agilent). Probes were selected to tile the proximal promoter regions of all known genes, that is, intervals roughly 2,000 bp upstream and 400 bp downstream of the transcription start sites of genes at 100-bp spacing. Extracted microarray intensities were processed and analyzed using the R software environment (16). Our model was adjusted for the four covariates identified by eigenR2 analysis (17) as those contributing more importantly to the variance: history of adversity (5.4%), postmortem interval (2.1%), substance use disorder comorbidity (1.7%), and age (1.7%). (See the online data supplement for complete descriptions of probe design and microarray analysis.)
Gene Expression Microarrays
Whole-genome gene expression data were obtained from gene expression microarrays previously generated by our group (18) using Affymetrix HU 133 plus2 GeneChip microarrays (Santa Clara). Methylation and expression data were compared in a subset of samples for which we also generated expression and methylation profiles (21 suicide completers and nine comparison subjects). Expression data were normalized as previously described (19). (See the online data supplement for complete descriptions of gene expression microarray analysis.)
Fluorescence-Assisted Cell Sorting and Microarray Validation
Neuronal nuclei were isolated from hippocampal tissue by fluorescence-assisted cell sorting using human anti-NeuN antibody conjugated to a fluorescent Alexa-488 marker as previously described (20). Nuclei were filtered and sorted on a FACSVantage SE system. Microarray validation was performed by EpiTYPER at the Genome Quebec Innovation Centre. Every sample used in the microarray experiments was used in the validation experiments. Results were analyzed by mixed-model analysis of variance (ANOVA) with group as a fixed factor and CpGs as a repeated measure, followed by least significant difference post hoc tests. The significance threshold was fixed at 0.05. (See the online data supplement for complete descriptions of fluorescence-assisted cell sorting and EpiTYPER analyses.)
Quantification of Gene Expression Using Quantitative Reverse Transcriptase Polymerase Chain Reaction
Total RNA was extracted from the same samples used in the microarray and validation experiments using the RNeasy lipid tissue extraction kit (Qiagen), followed by DNase I treatment, and cDNA conversion was performed using oligo(dT) primers. Gene expression was quantified using commercially available taqman primers and probe in an ABI7900HT (see Table S3 in the online data supplement; Applied Biosystems, Foster City, Calif.). Mean quantities from all samples were normalized to the reference genes β-actin and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) using commercially available primers and probe (see Table S3). Results were analyzed by univariate analysis of variance and adjusted for age, pH, postmortem interval, and RNA integrity number values.
Results
Probe Distribution
We used a genome-wide approach to map DNA methylation differences in the hippocampus of suicide completers relative to comparison subjects. There were no significant differences between groups in age, pH, and postmortem interval (see the online data supplement). In total, 330,600 probes were distributed over the promoter regions of 23,551 genes. The mean intensity was significantly different between the suicide and comparison groups at 366 probe locations after correction for multiple testing and significant covariates, including postmortem interval, history of adversity, substance use disorders, and age (Table 1; see also the online data supplement). From this list of differentially methylated regions, 273 (75%) probes indicated hypermethylation and 93 (25%) probes indicated hypomethylation in gene promoters of suicide completers.
| Methylation and Chromosome | Methylation Level | Corrected p | Log Fold Change | Gene | Distance From Transcription Start Site (bp) | Group |
|---|---|---|---|---|---|---|
| Hypermethylated in suicide completers | ||||||
| 7 | 1.0000 | 0.0032 | 1.1018 | MUC3A | 638 | Suicide |
| 17 | 0.9514 | 0.0037 | 1.4821 | KCNJ12 | 466 | Suicide |
| 17 | 0.9514 | 0.0037 | 1.2924 | KCNJ12 | 444 | Suicide |
| 17 | 0.9595 | 0.0037 | 1.0525 | KCNJ12 | 379 | Suicide |
| 14 | 1.0000 | 0.0037 | 1.3159 | SYNE2 | 500 | Suicide |
| 10 | 0.6210 | 0.0043 | 0.9216 | SYNPO2L | 298 | Suicide |
| 1 | 0.8826 | 0.0046 | 1.1937 | RPS6KA1 | 800 | Suicide |
| 1 | 0.8803 | 0.0046 | 0.7385 | RPS6KA1 | 253 | Suicide |
| 1 | 0.9050 | 0.0053 | 1.0831 | HIST2H2AB | 1,113 | Suicide |
| 1 | 0.9050 | 0.0053 | 0.9466 | HIST2H2AB | 1,074 | Suicide |
| 2 | 0.2822 | 0.0054 | 1.1356 | MIR10B | –96 | Suicide |
| 19 | 0.9058 | 0.0054 | 0.9742 | JUNB | 468 | Suicide |
| 11 | NA | 0.0057 | 0.9130 | SNX32 | 1,060 | Suicide |
| Hypomethylated in suicide completers | ||||||
| 14 | 0.6156 | 0.0031 | –0.8895 | SNORD114–14 | 950 | Comparison |
| 14 | 0.5840 | 0.0031 | –0.8581 | SNORD114–14 | 884 | Comparison |
| 12 | 0.0809 | 0.0032 | –0.9669 | CLEC12B | 708 | Comparison |
| 9 | 0.7779 | 0.0032 | –1.1428 | MAPKAP1 | –37 | Comparison |
| 3 | 0.6333 | 0.0040 | –1.2078 | PLCL2 | 320 | Comparison |
| Y | 0.8759 | 0.0071 | –1.2183 | AMELY | 211 | Comparison |
| Y | 0.5848 | 0.0073 | –0.9263 | NLGN4Y | 765 | Comparison |
| 9 | 0.6421 | 0.0073 | –0.9465 | OR13F1 | 1,177 | Comparison |
| 5 | 0.2893 | 0.0076 | –1.1632 | DNAH5 | 820 | Comparison |
| 6 | 0.6349 | 0.0077 | –0.9754 | CLIC5 | 545 | Comparison |
| 10 | NA | 0.0080 | –1.4672 | A1CF | 716 | Comparison |
| 15 | 0.8396 | 0.0111 | –0.8247 | TGM7 | 549 | Comparison |
| 15 | 1.0000 | 0.0111 | –0.9649 | TGM7 | 429 | Comparison |
Genomic Distribution and Relation With Expression
We first assessed the genomic distribution of differentially methylated probes between suicide completers and comparison subjects across the genome. We did not find any evidence for clusters of DNA methylation alterations, with probes being evenly distributed across the genome (Figure 1A). To estimate the statistical significance of this finding, we first computationally divided the genome into regions of 500 kb to 1 Mb, and found that only 20% of the differentially methylated probes were within 500 kb of each other. Permutation tests revealed that this proportion was in the expected range for the random distribution of 319 differentially methylated promoter regions.
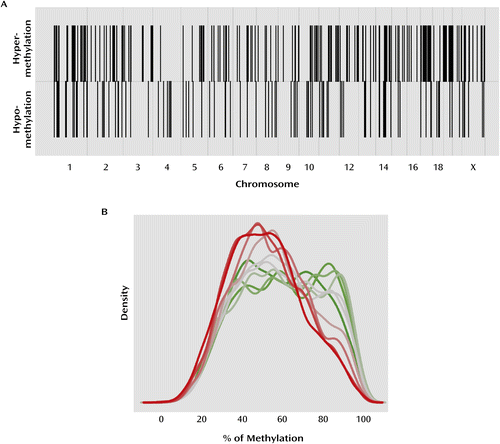
a Panel A shows the chromosomal distribution of differentially methylated probes in suicide completers relative to comparison subjects. The upper row shows sites that were hypermethylated relative to comparison subjects, and the lower row hypomethylated. Both hyper- and hypomethylated probes are evenly distributed across the genome. Panel B shows the relationship between gene expression and promoter DNA methylation across the genome. Genes were ordered by level of expression and split into 10 groups, from the lowest 10% to the highest 10%, with each line representing one of these groups (green=low expression; gray=medium expression; red=high expression). Genes with low expression typically have higher promoter methylation levels, whereas genes with high expression typically have lower promoter methylation levels (r=–0.14; p≤4.7×10−247).
DNA methylation in promoters has classically been associated with regulation of gene expression (4). The functional relationship between transcription and DNA methylation in promoters at a genome-wide level was assessed in our sample using gene expression data available from a substantial subgroup of the same tissue samples (Affymetrix HU 133 plus2). Mean expression levels across all samples in both groups were found to be significantly inversely correlated with mean promoter methylation levels across all samples (r=–0.14; p≤4.7×10−247) (Figure 1B; see also the online data supplement). More importantly, we found that mean gene expression and promoter methylation differences between suicide completers and comparison subjects were also inversely correlated. However, because differences are more subtle than variations in absolute levels of gene expression and promoter methylation, we first partitioned the genome into 1-Mb regions and calculated summaries of gene expression and promoter methylation differences between suicide completers and comparison subjects from all genes within each partition. These summarized gene expression and promoter methylation differences are inversely correlated across the genome (r=–0.19, p≤4.0×10−6).
Gene Ontology Analysis
The functional relevance of our findings was investigated through gene ontology analysis using the gene annotation algorithm in DAVID, version 6.7 (Database for Annotation, Visualization, and Integrated Discovery; http://david.abcc.ncifcrf.gov/), including every gene showing differential promoter methylation in suicide completers relative to comparison subjects (see Table S2 in the online data supplement). Results revealed an enrichment of methylation alterations in genes involved in, among other functions, cognitive processes, including learning, memory, and neuronal communication, such as synaptic transmission (Figure 2A; Table 2). In total, this functional cluster was composed of 33 genes divided into 10 ontological terms. The orphan nuclear receptor NR2E1 gene, the neuronal acetylcholine receptor subunit beta-2 (CHRNB2), the metabotropic glutamatergic receptor 7 (GRM7), and the dopamine beta-hydroxylase (DBH) genes were among the most differentially methylated genes within this cluster and were thus selected for follow-up validation (Figure 2).
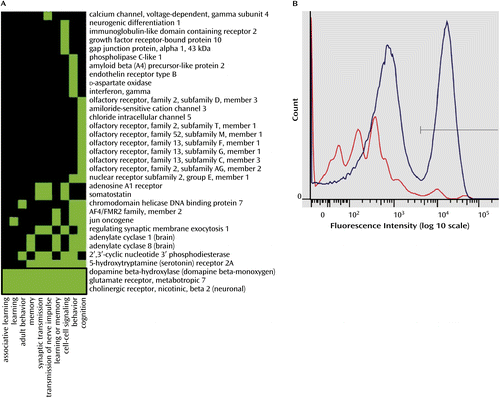
a The analysis was conducted using DAVID, version 6.7 (Database for Annotation, Visualization, and Integrated Discovery). In panel A, a green box indicates that the corresponding gene is known to play a role in the corresponding cellular function, and a black box indicates that no such role has been reported. Panel B shows that the total nuclei stained with NeuN generated a bimodal fluorescence intensity distribution. The intensity of fluorescence in the nonneuronal fraction (left blue peak) is less than in the neuronal fraction (right blue peak). The red line represents the intensity of fluorescence generated by the nonspecific fluorophore-conjugated secondary antibody without NeuN antibody.
| Cluster, Category, and Term | Count | % | p | Enrichment |
|---|---|---|---|---|
| Functional annotation cluster 1 (enrichment score, 1.50) | ||||
| Biological processes | ||||
| Memory | 6 | 1.97 | 3.6E-04 | 9.64 |
| Learning or memory | 8 | 2.63 | 2.0E-03 | 4.51 |
| Behavior | 16 | 5.26 | 8.1E-03 | 2.14 |
| Associative learning | 3 | 0.99 | 2.9E-02 | 11.05 |
| Adult behavior | 5 | 1.64 | 4.8E-02 | 3.64 |
| Learning | 4 | 1.32 | 7.0E-02 | 4.18 |
| Cognition | 20 | 6.58 | 1.3E-01 | 1.38 |
| Transmission of nerve impulse | 9 | 2.96 | 1.9E-01 | 1.61 |
| Synaptic transmission | 8 | 2.63 | 2.0E-01 | 1.68 |
| Cell-cell signaling | 12 | 3.95 | 3.6E-01 | 1.25 |
| Functional annotation cluster 2 (enrichment score, 1.15) | ||||
| Biological processes | ||||
| Regulation of carbohydrate metabolic process | 3 | 0.99 | 4.0E-02 | 9.39 |
| Regulation of cellular carbohydrate metabolic process | 3 | 0.99 | 4.0E-02 | 9.39 |
| Functional annotation cluster 3 (enrichment score, 1.14) | ||||
| Molecular function | ||||
| Phosphoric diester hydrolase activity | 5 | 1.64 | 4.2E-02 | 3.81 |
| Phosphoinositide phospholipase C activity | 3 | 0.99 | 5.7E-02 | 7.67 |
| Lipase activity | 5 | 1.64 | 7.1E-02 | 3.20 |
| Phospholipase C activity | 3 | 0.99 | 8.3E-02 | 6.19 |
| Phospholipase activity | 4 | 1.32 | 1.4E-01 | 3.08 |
Fluorescence-Assisted Cell Sorting
DNA methylation is cell-type specific, and neurons are likely to exhibit DNA methylation patterns different from those of nonneuronal cell types (21). To determine whether methylation differences found in the arrays were more pronounced in the neuronal or the nonneuronal cell fraction, we performed the validation of our microarray data on DNA from both neuronal and nonneuronal cell populations isolated by fluorescence-assisted cell sorting. Fluorophore-conjugated anti-NeuN antibody was used to target neuronal nuclei in hippocampal tissue, and a bimodal distribution of neuronal and nonneuronal nuclei was obtained (Figure 2B).
Experimental Validation
Microarray validation was performed on four genes represented in the first functional cluster (NR2E1, CHRNB2, GRM7, and DBH) on both neuronal and nonneuronal DNA. The methylation patterns and expression profiles generated for these four genes through microarray analysis were successfully confirmed by EpiTYPER. Promoter hypermethylation was found more frequently in the promoters of NR2E1, CHRNB2, GRM7, and DBH in neuronal nuclei as compared with nonneuronal nuclei. Our results also suggest the presence of different methylation signatures in neuronal and nonneuronal cell populations. In addition, we quantified the expression of genes that were selected for validation. In accordance with the overall inverse correlation between promoter methylation and gene expression reported in this study, our results suggest that DNA methylation in the promoters of validated genes is associated with differential expression in the brain samples of suicide completers. Here we describe only NR2E1 results; complete results on CHRNB2, GRM7, and DBH methylation and expression data are provided in Figure S4 in the online data supplement.
We measured DNA methylation levels in a 466-bp region in the NR2E1 promoter (chr6:108,488,081 to 108,488,545) including 28 CpGs. In the neuronal fraction, we found a significant hypermethylation in the suicide completers relative to the comparison subjects (mixed-model ANOVA: F=13.24, df=1, 165, p<0.0005) (Figure 3A). Post hoc analyses revealed significant hypermethylation in suicide completers relative to comparison subjects at CpG sites 15 (p<0.005), 22, 23 (p<0.05), 24 (p<0.05), and 28 (p<0.005) (Figure 3E).

a Total percent of methylation in NR2E1 promoter in the neuronal cell fraction (panel A; suicide completers, N=35; comparison subjects, N=14) and the nonneuronal cell fraction (panel B; suicide completers, N=46; comparison subjects, N=16). Panel C shows the relative expression of NR2E1 in the hippocampus (suicide completers, N=46; comparison subjects, N=15). In panel D, the graph shows an inverse correlation between NR2E1 mean total % of methylation and NR2E1 relative expression. The graphs in panels E and F show the individual CpG methylation levels in the promoter of NR2E1 in the neuronal and nonneuronal cell fractions, respectively. Values are mean % of methylation, and error bars indicate standard error of the mean. *p<0.05; †p<0.1.
Similar results were obtained in the nonneuronal cell fraction. Indeed, a significant hypermethylation was found in the suicide completers relative to comparison subjects (F=10.53, df=1, 271, p<0.005) (Figure 3B). Post hoc analyses in the nonneuronal cell fraction revealed significant hypermethylation at CpG sites 18 (p<0.01), 19 (p<0.05), 20 (p<0.005), 21 (p<0.005), and 25 (p<0.005) in the suicide completers relative to the comparison subjects (Figure 3F). Overall, our data show that a region of 300 bp within the NR2E1 promoter, and including 14 CpGs, is strongly hypermethylated in neurons and nonneuronal cells of suicide completers relative to comparison subjects.
The hypermethylation found in NR2E1 promoter was also associated with a significantly lower expression of the NR2E1 gene in the hippocampus of suicide completers relative to comparison subjects (F=5.10, df=1, 56, p<0.05) (Figure 3C) that was inversely correlated with overall mean DNA methylation levels in the neuronal cell fraction (r=–0.308, p=0.05) (Figure 3D). We did not observe a similar interaction in the nonneuronal cell fraction. Together, these data suggest that DNA methylation within this region of NR2E1 promoter may have a regulatory role on NR2E1 expression.
Discussion
We examined DNA methylation patterns in the hippocampus of suicide completers and nonpsychiatric sudden-death comparison subjects at the proximal promoter regions of all known genes. In accordance with previous gene candidate studies (12), our results suggest that promoter DNA methylation levels are significantly greater for several genes in suicide completers relative to comparison subjects. In addition, our results reveal the existence of a significant number of hypomethylated sequences in the promoters of suicide completers, suggesting that DNA methylation patterns are altered in the brains of suicide completers, with both hyper- and hypomethylation found in gene promoters across the genome.
DNA methylation is a major regulator of gene expression (4). In agreement with previous findings in brain tissue (12), our results show that overall in the genome, gene expression is negatively correlated with promoter methylation. This observation is also supported by our expression data on validated genes. Differentially methylated regions were found evenly distributed across the genome, an observation that has also been reported in the blood of patients with posttraumatic stress disorder and in brain tissue of psychotic and bipolar subjects (22, 23), suggesting that epigenetic changes in psychiatric disorders may not be limited to a short list of specific gene candidates but may affect multiple functional gene networks in multiple chromosomal regions.
Suicide is a complex problem that most likely results from multiple pathological pathways (24). Epigenetic alterations are expected to increase vulnerability to suicide by interfering with normal gene expression patterns, leading to neurobiological abnormalities associated with the development of specific emotional and behavioral phenotypes (25, 26) as well as cognitive impairments (27, 28). The biological functions identified by our ontological analyses, as the most significantly enriched in genes with differential promoter methylation, are related to behavioral and cognitive processes such as learning and memory. This implies that the epigenetic regulation of these processes in the brain may be altered in suicide completers, leading to the dysregulation of cognitive processes.
We targeted four genes known to be involved, whether directly or indirectly, in the regulation of the cellular processes of learning, memory, and behavior (29–33). NR2E1 encodes a brain-specific orphan nuclear receptor acting as a transcriptional repressor (33), and GRM7 codes for the G-protein coupled metabotropic glutamate receptor subunit 7 (34). CHRNB2 encodes a brain-specific subunit (β2) of the ligand-gated ionotropic nicotinic acetylcholine receptor family (35, 36), and DBH codes for a catecholamine-synthetic membrane-bound enzyme responsible for the synthesis of norepinephrine (37).
Our results revealed that both NR2E1 and GRM7 promoter methylation levels were associated with lower gene expression in the hippocampus of suicide completers. Interestingly, NR2E1 knockout mice exhibit severely aggressive and impulsive behaviors, blunted anxiety, fear conditioning, learning and memory deficits, and reduced mating (32). Similarly, GRM7−/− mice display a marked reduction in fear-mediated freezing responses (38) and short-term working memory deficits in the four- and eight-arm maze task (39). These behavioral phenotypes parallel many behavioral traits in humans that are regarded as risk factors for suicidal behavior. For instance, developmental trajectories characterized by high anxiety traits and externalizing behaviors (impulsivity, aggression) are predictive of suicide (40). It is possible that changes in DNA methylation within promoters of a subset of genes regulating behavioral traits may lead to regulatory changes in the establishment of stable emotional and behavioral trajectories and associate with higher levels of anxiety, aggression, and impulsivity, which may in turn increase vulnerability to suicide.
Vulnerability to suicide has also been associated with several cognitive deficits (28). Interestingly, in addition to NR2E1 and GRM7, we found differential promoter methylation in CHRNB2 and DBH, two genes that have been directly or indirectly associated with learning and memory formation by modulating long-term potentiation or long-term depression (29, 31). Chronic, but not acute, pretreatment with nicotine reverses the effects of chronic psychosocial stress on long-term potentiation and long-term depression in rats (41, 42). Consistently with the cognitive deficits observed in individuals with a past history of suicidal acts, these deficits seem to be in close relationship with the capacity to respond effectively to stressful situations. Indeed, individuals with altered hypothalamic-pituitary-adrenal axis reactivity fail to improve in retest of executive function (43) and show more decision-making impairments (44) following stress. Together, our results showing differential promoter DNA methylation associated with changes in the expression of genes involved in the cellular processes of learning and memory are consistent with current theoretical models of the neurobiology of suicidal behavior, and thus it is possible that these alterations are etiologically related to suicide (45).
On the other hand, the role of DBH in the hippocampus may rather be indirect through its role in the synthesis of norepinephrine. Indeed, norepinephrine is suspected to enhance contextual fear memory and long-term potentiation in the hippocampus through phosphorylation of GluR1 subtypes, facilitating AMPA trafficking at synaptic sites (46). However, our data show no difference in DBH expression between suicide completers and comparison subjects, suggesting that other mechanisms regulating expression, such as histone modifications, may be involved. More research is needed to elucidate the complex relationship between DNA methylation and histone modifications.
Concordant with their expression patterns, the hypermethylation in the promoter of NR2E1, CHRNB2, GRM7, and DBH was almost specific to the neuronal cell fraction. Indeed, CHRNB2, GRM7, and DBH are expressed only in neurons (37, 47, 48), whereas NR2E1 expression is not restricted to neurons. Our data suggest that the hypermethylation found within the neuronal cell fraction may be responsible for the lower expression of NR2E1, GRM7, and CHRNB2 genes in the hippocampus of suicide completers.
However, we found significant hypomethylation in CHRNB2 and GRM7 promoters in the nonneuronal cell fractions, suggesting that the same genes could be differently affected in different cell types, with cellular consequences proper to each cell type. This is consistent with the fact that different tissues (49) and cell types within the same tissue (21, 50) exhibit different DNA methylation patterns. However, it is hard to speculate on the functional impact of these findings, especially given that these observations were made on a highly heterogeneous cell population composed of different nonneuronal cell types and that these genes are expressed primarily in neurons.
Our results are concordant with results we recently published on the epigenetic effects of early-life adversity in the brain (51). While early-life adversity was also associated with a genome-wide epigenetic reprogramming, the differentially methylated genes observed in the two studies were different and are known to be involved in different functional pathways. Thus, the data in the present study expand our understanding of the role of epigenetic mechanisms in the brains of suicide completers and provide future avenues of investigation in the study of molecular processes in the brains of suicide completers.
The data we present here have been adjusted for covariates susceptible to influencing DNA methylation and expression (history of adversity, postmortem interval, substance use disorder comorbidity, and age). It is known, however, that medication alters DNA methylation levels both in the brain and in peripheral tissue (52, 53). Although our analyses suggest that medication may not have significantly affected DNA methylation in our study, more research is needed to investigate the role of medication in DNA methylation in the human brain.
One important consideration regarding our study is the method used to isolate methylated DNA. MeDIP is a highly sensitive method for the enrichment of methylated genomic DNA (15) that is not limited by sequence context of methylation-specific enzymes and does not require extensive bisulfite treatment. Although MeDIP enriches DNA sequences with both low and high CpG density, it is more sensitive to methylation in CpG-rich regions and CpG islands (54). The extent of promoters’ CpG enrichment varies throughout the genome (55), and CpG density within promoters has been associated with different DNA methylation levels. Generally, CpG-rich promoters have been associated with low DNA methylation levels, while low-CpG density promoters have been shown to be more methylated (55, 56). Thus, it is possible that our results represent only a subset of even larger methylation changes taking place throughout the genome, given that some differences in gene promoters with low CpG density may not be well represented.
In addition, it has been suggested that the 5′ methylcytosine antibody may capture, to a certain extent, non-CpG methylation (54), which has classically been reported in embryonic and induced pluripotent stem cells (57–59), but also recently in mouse brains (60). It is unlikely, however, that this happened in our study. Indeed, our analyses were focused on gene promoters, and non-CpG DNA methylation has been reported mainly in gene bodies (57). However, more research is needed to address this question.
In summary, our results show DNA methylation differences in gene promoter regions throughout the genome in the hippocampus associated with suicide. These changes in methylation levels are in genes involved in the regulation of behavioral and cognitive processes that have been shown to be altered in individuals with suicidal behaviors. Thus, our findings suggest that such mechanisms may, in individuals with particular behavioral, molecular, and cellular predispositions, increase susceptibility to suicide.
1 World Health Organization: Suicide Prevention (SUPRE). http://www.who.int/mental_health/prevention/suicide/suicideprevent/en/Google Scholar
2 : Suicide neurobiology. Prog Neurobiol 2009; 89:315–333Crossref, Medline, Google Scholar
3 : Personality traits as prospective predictors of suicide attempts. Acta Psychiatr Scand 2009; 120:222–229Crossref, Medline, Google Scholar
4 : Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 2006; 31:89–97Crossref, Medline, Google Scholar
5 : Chromatin modifications and their function. Cell 2007; 128:693–705Crossref, Medline, Google Scholar
6 : The early life social environment and DNA methylation: DNA methylation mediating the long-term impact of social environments early in life. Epigenetics 2011; 6:971–978Crossref, Medline, Google Scholar
7 : Alternative splicing, methylation state, and expression profile of tropomyosin-related kinase B in the frontal cortex of suicide completers. Arch Gen Psychiatry 2009; 66:22–32Crossref, Medline, Google Scholar
8 : Increased BDNF promoter methylation in the Wernicke area of suicide subjects. Arch Gen Psychiatry 2010; 67:258–267Crossref, Medline, Google Scholar
9 : GABAA receptor promoter hypermethylation in suicide brain: implications for the involvement of epigenetic processes. Biol Psychiatry 2008; 64:645–652Crossref, Medline, Google Scholar
10 : Differential glucocorticoid receptor exon 1(B), 1(C), and 1(H) expression and methylation in suicide completers with a history of childhood abuse. Biol Psychiatry 2012; 72:41–48Crossref, Medline, Google Scholar
11 : Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci 2009; 12:342–348Crossref, Medline, Google Scholar
12 : The epigenetics of suicide: explaining the biological effects of early life environmental adversity. Arch Suicide Res 2010; 14:291–310Crossref, Medline, Google Scholar
13 : The Human Brain: An Introduction to Its Functional Neuroanatomy, 5th ed. St Louis, MO, Mosby-Year Book, 2002Google Scholar
14 : Linkage disequilibrium analysis in young populations: pseudo-vitamin D-deficiency rickets and the founder effect in French Canadians. Am J Hum Genet 1996; 59:633–643Medline, Google Scholar
15 : Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet 2006; 38:149–153Crossref, Medline, Google Scholar
16
17 : Eigen-R2 for dissecting variation in high-dimensional studies. Bioinformatics 2008; 24:2260–2262Crossref, Medline, Google Scholar
18 : Profiling brain expression of the spermidine/spermine N1-acetyltransferase 1 (SAT1) gene in suicide. Am J Med Genet B Neuropsychiatr Genet 2009; 150B:934–943Crossref, Medline, Google Scholar
19 : Patterns of gene expression in the limbic system of suicides with and without major depression. Mol Psychiatry 2007; 12:640–655Crossref, Medline, Google Scholar
20 : Neuronal nuclei isolation from human postmortem brain tissue. J Vis Exp 2008 (doi: 10.3791/914)Crossref, Google Scholar
21 : Neurons show distinctive DNA methylation profile and higher interindividual variations compared with non-neurons. Genome Res 2011; 21:688–696Crossref, Medline, Google Scholar
22 : Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet 2008; 82:696–711Crossref, Medline, Google Scholar
23 : Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc Natl Acad Sci USA 2010; 107:9470–9475Crossref, Medline, Google Scholar
24 : The neurodevelopmental origins of suicidal behavior. Trends Neurosci 2012; 35:14–23Crossref, Medline, Google Scholar
25 : Broad and narrow personality traits as markers of one-time and repeated suicide attempts: a population-based study. BMC Psychiatry 2008; 8:15Crossref, Medline, Google Scholar
26 : Personality traits as correlates of suicidal ideation, suicide attempts, and suicide completions: a systematic review. Acta Psychiatr Scand 2006; 113:180–206Crossref, Medline, Google Scholar
27 : Impaired decision making in suicide attempters. Am J Psychiatry 2005; 162:304–310Link, Google Scholar
28 : The suicidal mind and brain: a review of neuropsychological and neuroimaging studies. World J Biol Psychiatry 2011; 12:319–339Crossref, Medline, Google Scholar
29 : The cholinergic system and hippocampal plasticity. Behav Brain Res 2011; 221:505–514Crossref, Medline, Google Scholar
30 : Metabotropic glutamate receptor 7: at the interface of cognition and emotion. Eur J Pharmacol 2010; 639:123–131Crossref, Medline, Google Scholar
31 : Emotional enhancement of memory: how norepinephrine enables synaptic plasticity. Mol Brain 2010; 3:15Crossref, Medline, Google Scholar
32 : A tale of tailless. Dev Neurosci 2011; 33:1–13Crossref, Medline, Google Scholar
33 : Nuclear receptors in stem cells and their therapeutic potential. Adv Drug Deliv Rev 2010; 62:1299–1306Crossref, Medline, Google Scholar
34 : Metabotropic glutamate receptors as novel targets for anxiety and stress disorders. Nat Rev Drug Discov 2005; 4:131–144Crossref, Medline, Google Scholar
35 : Relating neuronal nicotinic acetylcholine receptor subtypes defined by subunit composition and channel function. Mol Pharmacol 2003; 63:311–324Crossref, Medline, Google Scholar
36 : Structure and gating mechanism of the acetylcholine receptor pore. Nature 2003; 423:949–955Crossref, Medline, Google Scholar
37 : Dopamine beta-hydroxylase of adrenal chromaffin granules: structure and function. Annu Rev Biochem 1988; 57:551–592Crossref, Medline, Google Scholar
38 : Metabotropic glutamate receptor subtype 7 ablation causes deficit in fear response and conditioned taste aversion. J Neurosci 1999; 19:955–963Crossref, Medline, Google Scholar
39 : Lack of the metabotropic glutamate receptor subtype 7 selectively impairs short-term working memory but not long-term memory. Behav Brain Res 2004; 154:473–481Crossref, Medline, Google Scholar
40 : Childhood trajectories of anxiousness and disruptiveness as predictors of suicide attempts. Arch Pediatr Adolesc Med 2008; 162:1015–1021Crossref, Medline, Google Scholar
41 : Chronic but not acute nicotine treatment reverses stress-induced impairment of LTP in anesthetized rats. Brain Res 2006; 1097:78–84Crossref, Medline, Google Scholar
42 : Nicotine prevents stress-induced enhancement of long-term depression in hippocampal area CA1: electrophysiological and molecular studies. J Neurosci Res 2006; 83:309–317Crossref, Medline, Google Scholar
43 : Dysregulation of the sympathetic nervous system, hypothalamic-pituitary-adrenal axis, and executive function in individuals at risk for suicide. J Psychiatry Neurosci 2010; 35:399–408Crossref, Medline, Google Scholar
44 : Stress and decision-making in humans: performance is related to cortisol reactivity, albeit differently in men and women. Psychoneuroendocrinology 2009; 34:1449–1458Crossref, Medline, Google Scholar
45 : The neurobiology of depression: inroads to treatment and new drug discovery. J Clin Psychiatry 2005; 66(Suppl 7):5–13Crossref, Medline, Google Scholar
46 : Emotion enhances learning via norepinephrine regulation of AMPA-receptor trafficking. Cell 2007; 131:160–173Crossref, Medline, Google Scholar
47 : Mouse strain-specific nicotinic acetylcholine receptor expression by inhibitory interneurons and astrocytes in the dorsal hippocampus. J Comp Neurol 2004; 468:334–346Crossref, Medline, Google Scholar
48 : Molecular characterization of a new metabotropic glutamate receptor mGluR7 coupled to inhibitory cyclic AMP signal transduction. J Biol Chem 1994; 269:1231–1236Medline, Google Scholar
49 : DNA methylation signatures within the human brain. Am J Hum Genet 2007; 81:1304–1315Crossref, Medline, Google Scholar
50 : Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res 2011; 21:1074–1086Crossref, Medline, Google Scholar
51 : Genome-wide epigenetic regulation by early-life trauma. Arch Gen Psychiatry 2012; 69:722–731Crossref, Medline, Google Scholar
52 : Resilience to social stress coincides with functional DNA methylation of the Crf gene in adult mice. Nat Neurosci 2010; 13:1351–1353Crossref, Medline, Google Scholar
53 : Epigenetic regulation of BDNF expression according to antidepressant response. Mol Psychiatry (Epub ahead of print, May 1, 2012)Google Scholar
54 : Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol 2010; 28:1097–1105Crossref, Medline, Google Scholar
55 : Distribution, silencing potential, and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 2007; 39:457–466Crossref, Medline, Google Scholar
56 : Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008; 454:766–770Crossref, Medline, Google Scholar
57 : Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009; 462:315–322Crossref, Medline, Google Scholar
58 : Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci USA 2000; 97:5237–5242Crossref, Medline, Google Scholar
59 : Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet 2011; 7:e1002389Crossref, Medline, Google Scholar
60 : Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell 2012; 148:816–831Crossref, Medline, Google Scholar



