A Schizophrenia Gene Locus on Chromosome 17q21 in a New Set of Families of Mexican and Central American Ancestry: Evidence From the NIMH Genetics of Schizophrenia in Latino Populations Study
Abstract
Objective: The present study investigated a new set of families of Latin American ancestry in order to detect the location of genes predisposing to schizophrenia and related psychotic disorders. Method: A genome-wide scan was performed for 175 newly recruited families with at least two siblings suffering from a psychotic disorder. Best-estimate consensus procedures were used to arrive at diagnoses, and nonparametric allele-sharing statistics were calculated to detect linkage. Results: Genome-wide significant evidence for linkage for the phenotype of DSM-IV schizophrenia or schizoaffective disorder was found in a region on chromosome 17q21 (lod score, 3.33). A region on chromosome 15q22-23 showed suggestive evidence of linkage with this same phenotype (lod score, 2.11). Analyses using a broader model (any psychosis) yielded evidence of suggestive linkage for the 17q21 region only, and no region achieved genome-wide significance of linkage. Conclusions: The new set of 175 families of Mexican and Central American ancestry delineates two new loci likely to harbor predisposition genes for schizophrenia and schizoaffective disorder. The region with the strongest support for linkage in this sample, 17q21, has been implicated in meta-analyses of schizophrenia genome screens, but the authors found no previous reports of it as a locus for schizophrenia in specific population- or family-based studies, and it may represent the location of a schizophrenia predisposition gene (or genes) of special relevance in Mexican and Central American populations.
Genome-wide linkage scans of families with multiple cases of schizophrenia have been a valuable tool in identifying genes that contribute to this disorder in the general population (1 – 5) . Beginning in the mid-1990s, genome-wide linkage screens of multiplex schizophrenia families have resulted in identification of several chromosomal regions that displayed significant linkage results in distinct populations (4 , 6 – 8) . Although replication of results has been difficult, as is to be expected in the study of illnesses that are genetically complex, initial localization of schizophrenia predisposition genes to chromosomes 1, 6, 8, and 13 has led to eventual determination of specific genes that are associated with schizophrenia (9) . Previously, we reported the results of phase 1 of the National Institute of Mental Health (NIMH) Genetics of Schizophrenia in Latino Populations Study, which revealed the main loci for psychosis genes in 99 families of Mexican and Central American origin (8) . Here we report the results of phase 2 of this study, in which 175 new multiplex families of Mexican and Central American origin were analyzed in a genome-wide screen. This new set of families was recruited from the southwestern United States, Mexico, and Central America. In this new sample, we report evidence of genome-wide significant linkage for the phenotype of schizophrenia to chromosome 17. This is a chromosomal location that has not been significantly linked to schizophrenia in any previous population study of which we are aware, although it has been suggested as a potential locus for schizophrenia in meta-analyses.
Method
Subjects
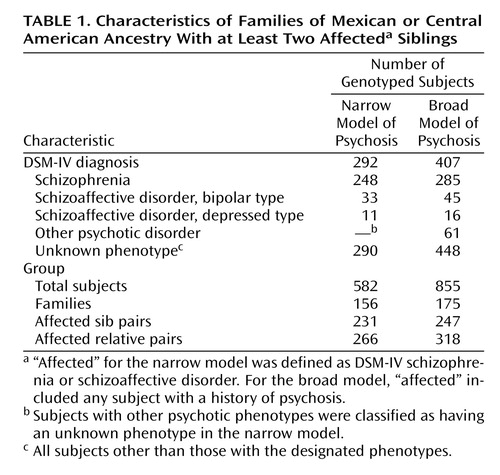
Families were recruited from sites throughout the southwestern United States, Mexico, and Central America. Each family had at least two siblings who had been previously diagnosed with either schizophrenia or schizoaffective disorder (according to inpatient or outpatient records). Assessment of affected subjects was conducted as described previously (8) . In brief, clinical material was collected for each affected individual, including a direct interview by means of the Diagnostic Interview for Genetic Studies (version 2.0) (10) , an interview with a close relative using the Family Interview for Genetic Studies (11) , and any available hospitalization or treatment records. Assessments with the Diagnostic Interview for Genetic Studies were all conducted in the subject’s preferred language, by a psychiatrist who had participated in reliability training exercises with the research consortium. Final DSM-IV lifetime diagnoses were assigned by a consensus best-estimate team, as previously described (8) . This team also provided consensus information on lifetime history of psychosis, as defined by any of criteria A 1–4 of the DSM-IV criteria for schizophrenia.
Table 1 lists the total number of families included in this current analysis, as well as the number of affected relative pairs under the broad and narrow models, and the diagnoses of the subjects studied. No families from the previous sample (8) were included in this current analysis. After consensus diagnoses were completed, the number of affected sibling pairs was 247 for the broad analysis (plus 71 other types of affected relative pairs) and 231 for the narrow analysis (plus 35 additional types of affected relative pairs). This group of genotyped affected sibling pairs is more than twice the size of our previously studied set of 99 families, which had 81 and 70 fully genotyped affected sib pairs, respectively, for the broad and narrow models.
All subjects provided written informed consent after the study procedures were explained to them.
Genotyping
Blood samples were drawn by using sterile technique and processed at the NIMH Genetics Initiative cell repository at Rutgers University. DNA for 855 individuals, all from this new set of 175 families, was processed and shipped to the Center for Inherited Disease Research at Johns Hopkins University for genotyping. Each individual’s DNA was genotyped there for 385 single tandem repeat markers, covering the genome at approximately 10-centimorgan (10-cM) intervals. There was no overlap of the subjects or families genotyped for the present analyses and those reported in our previous study of Latin American families (8) .
Error Checking
The genotype data were checked for inconsistencies with pedigree data by using Preswalk, a version of Simwalk2 (12) that accepts data in PEDSYS database format (13) . Simwalk2 uses a Markov-chain Monte Carlo procedure to determine the likelihood of the data set with the assumption that genotyping errors cause unlikely recombinations between loci. This program also detects Mendelian errors caused by pedigree problems such as nonpaternity and detects monozygotic twins. In addition, the linkage analysis software MERLIN checks for pedigree errors before initiating each analysis (14) .
Linkage Analysis
We analyzed two related phenotypes for linkage, following the same protocol we used in our previous study of the first 99 families in the NIMH Genetics of Schizophrenia in Latino Populations study (8) . Our broad model considered any subject with a consensus lifetime history of psychosis as affected, and it considered all other subjects to have an unknown phenotype. A subject was considered to have a lifetime history of psychosis if he or she was considered by the best-estimate consensus team to have met at least one of the first four A criteria for schizophrenia in DSM-IV. In the present sample, the DSM-IV consensus diagnoses of the subjects who had a consensus lifetime history of psychosis were as follows: schizophrenia (N=285), schizoaffective disorder, bipolar (N=45), schizoaffective disorder, depressed (N=16), major depressive disorder with psychosis (N=14), bipolar type I disorder with psychosis (N=5) and psychotic disorder not otherwise specified (N=42). A narrow model was also used, in which only subjects with a consensus DSM-IV diagnosis of schizophrenia or schizoaffective disorder (either type) were considered affected and all other subjects were listed as having an unknown phenotype. For the current phase 2 analyses, there were 175 new families under the broad model and 156 new multiplex families under the narrow model. Nonparametric linkage analyses were performed by using the MERLIN computer program (14) . A sib pair model was used in which only affected individuals were considered; unaffected individuals were represented as having unknown diagnostic status. MERLIN conducts multipoint linkage analysis by means of the Kong and Cox linear model (15) .
To assess the genome-wide significance of findings of linkage, 10,000 gene-dropping simulations were carried out for each model by using MERLIN with the broad and narrow phenotypes. The simulation process assigns random chromosomes to founders by using genotype frequencies estimated by means of maximum likelihood. The process assigns chromosomes to individuals in the pedigree while taking into account their relationships to each other and recombination frequencies. This system allows for the computation of false positive rates since it retains the original pattern of relationships between relatives and missing data and the simulated data are unlinked to the trait. The method of Feingold et al. (16) was used to obtain p values for lod scores (logarithms of the odds ratios for linkage). Using the criteria established by Lander and Kruglyak (17) , we defined suggestive evidence of linkage as any lod score that would be expected to occur no more than once per genome scan by chance. According to our simulations, this would be equivalent to a lod score of 1.71 or higher.
Results
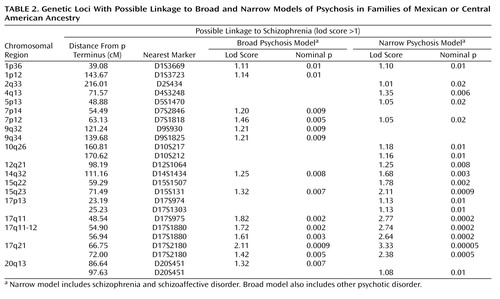
In this new set of families, evidence of suggestive or significant linkage (lod score ≥1.71) was found at two loci, as shown in Table 2 . Complete plots of the multipoint linkage scores for both the broad and narrow psychosis models are displayed in Figure 1 . For the broad phenotype ( Table 2 ), eight loci had lod scores greater than 1.0 (corresponding to a nominal p value of 0.01), on chromosomes 1 (two loci), 7, 9, 14, 15, 17, and 20, with the highest peak on chromosome 17. The peak on chromosome 17 meets the criteria of suggestive linkage, as defined by our simulation analysis and according to the standard of Lander and Kruglyak (lod scores expected to happen by chance once per genome screen) (17) . None of these loci met the genome-wide criteria for linkage in our sample. For the narrow model ( Table 2 ), 11 loci had lod scores greater than 1.0, on chromosomes 1, 2, 4, 5, 7, 10, 12, 14, 15, 17, and 20. The peak multipoint lod score was on chromosome 17q21. This evidence of linkage is close to the level proposed by Lander and Kruglyak (17) for significant genome-wide linkage in a sib pair study (lod score=3.6, p=2.0×10 –5 ) and meets genome-wide significant linkage according to our simulations (described in the Method section). Our simulations indicated that a lod score of 3.33 or higher would be expected to occur 2.09 times per 100 genome scans (individual lod scores at or above this level occurred in 209 individual marker simulations out of 10,000 whole genome screen simulations with the given data structure).
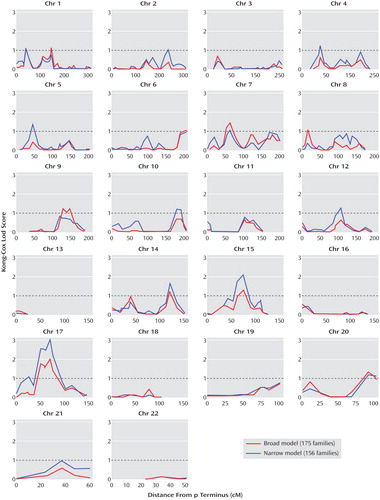
a The narrow phenotype includes schizophrenia and schizoaffective disorder. The broad phenotype includes any history of psychosis. A lod score of 1.0 is equivalent to a nominal p of 0.01.
Besides the peak on chromosome 17 (which technically has lod scores greater than 2.0 spanning from 17q11 to 17q21), only one other region attained evidence for linkage with a lod score greater than 2.0 ( Table 2 ). This locus was also for the narrow model and was located in the chromosome 15q22-24 region, with a peak lod score of 2.11, which meets the criteria for suggestive evidence of linkage in the present study.
Discussion
The results of the present study reveal several new loci of interest in the search for genes that predispose to schizophrenia in the Latino population. Although all of the loci listed in Table 2 are worthy of following up in future analyses and studies, the current sample highlights two key areas of interest, at 15q22-24 and at 17q21. The first locus shows suggestive evidence of linkage to the phenotype of schizophrenia and schizoaffective disorder, with the latter locus meeting genome-wide significance criteria in this sample. Although primary evidence of linkage at these sites has not been reported in any other specific studies of schizophrenia to our knowledge, both loci have been suggested as possible loci for this disease in meta-analyses (to be discussed). For both loci, broadening of the phenotype in the current study to include all cases of psychosis in these families did not increase the evidence of linkage.
This current set of 175 families is distinct from the set of 99 families previously studied (8) , although both sets of families were recruited because they had Mexican or Central American ancestry, and assessment, diagnosis, and the phenotypes analyzed were the same. In the first 99 families, genome-wide significant linkage for the broad phenotype of psychosis was demonstrated on chromosome 1p (peak at 37 cM from the p terminus). In this new set of families, this locus showed confirmatory evidence of linkage (both broad and narrow models had lod scores of 1.1 with a nominal p value of 0.012, at 39 cM from the p terminus). Although the second set of families (current study) yielded strong evidence of linkage at 17q21, this had not been suggested as a locus in the first set of families. Our results suggest the potential of stochastic differences between the two samples (i.e., by chance, the first set of families contained more families in which a gene or genes in the chromosome 1p region were responsible for the enhanced risk of psychosis, whereas the second set contained more families with a predisposition gene or genes in the chromosome 17q21 region). This could be due in part to geographical differences in the sampling; for instance, 16.9% of the subjects in the first study were from Central America, whereas 25.7% of those in the second study were from Central America (47.5% and 42.2%, respectively, were from Mexico, and 35.6% and 32.1% were from the southwestern United States). However, our previous analyses of population structure in the southwestern United States, Mexico, and Central America suggest that these populations share common ancestry and that there is more genetic heterogeneity within each geographical region than between regions (18) .
The fact that the 17q21 locus did not show evidence of linkage in the previous study (first sample) may attest to the fortuitous nature of linkage findings in moderate-sized samples. Definitively identifying genes for complex disorders, especially in outbred populations, may require very large samples of sibling pairs (19) . For oligogenic diseases, it is much more likely that genetic loci will be identified in initial screens than that they will be identified in a replication sample, and replication samples may need to be five to six times larger than the original sample to adequately test for replication (20) . Joint studies, in which all samples from our previous and current linkage studies are combined, were not possible to perform at this time, as allele calling and several markers were different in the two sets of families; future combined analyses of these families may thus yield additional loci of interest.
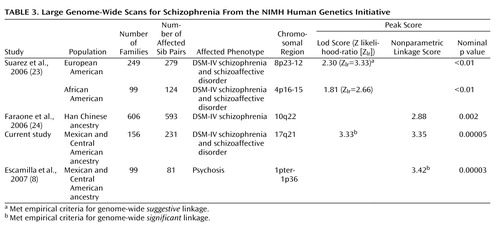
The present study, with 175 families, is one of the larger published studies using affected sibling pairs to study schizophrenia. In a summary of schizophrenia linkage screens, the average number of families in 20 linkage studies was 60 (and the average number of affected individuals per study was 147), with only two studies having as many as 170 families (21) . As part of the NIMH Human Genetics Initiative (22) , two other groups have studied large numbers of families by using a focus on affected sibling pairs, a narrow diagnosis (schizophrenia and schizoaffective disorder), and similar diagnostic procedures (Diagnostic Interview for Genetic Studies, Family Interview for Genetic Studies, consensus process, and DSM-IV diagnoses). Table 3 lists the resultant genome scans from these two groups, juxtaposed with the two sets of families from our analyses, and the resulting genomic loci identified, with results broken down by the ethnic origin of the families. One study reported on genome screens for 249 European-ancestry American families and 99 African-ancestry American families (23) , whereas another group reported on 606 Han Chinese families drawn from the Taiwanese population (24) . As in our study, the study on European- and African-ancestry American families designated an affected phenotype as DSM-IV schizophrenia or schizoaffective disorder (25) . The study from the Han Chinese population used a more restrictive diagnosis of DSM-IV schizophrenia for their analysis of affected persons, but the authors noted that when their phenotype was broadened to include schizoaffective disorder, “the results were virtually identical” to those they presented (24) . It is interesting that these studies, despite using similar methods, highlight different chromosomal regions of most significant linkage and that the linkage scores do not correlate directly with the number of affected sib pairs studied (the highest lod score comes from the current set of 231 Latino sib pairs, based on a narrow model, while the Han Chinese sample, which had the highest number of affected sib pairs, gave one of the lowest peak linkage scores of any of the studies).
Chromosome 17q21
In the current study, our strongest evidence for the location of a schizophrenia predisposition gene (or genes) was in the 17q21 region. In a meta-analysis of 20 genome scans for schizophrenia, performed by Lewis et al. in 2003 (21) , this region was implicated as one of a few that they felt could be possible loci for schizophrenia (one of 19 loci with “nominally significant” probability values), although only one locus (on chromosome 2) in their meta-analysis showed significant linkage. Lewis et al. noted that the 17q locus was highlighted by the meta-analyses even though it had not been previously implicated in individual studies. In searching the literature for evidence of linkage to this region, we identified only one specific study that implicated this region. In that study (25) a linkage analysis was performed to identify genes moderating the age at onset of schizophrenia, and marker 17S787, on chromosome 17q, gave the peak lod score for the genome screen.
Since the linkage examined is to chromosomal regions, rather than to specific genes, follow-up studies of this region will need to carefully screen many genes. Nevertheless, it is interesting that this locus has been implicated in several other CNS diseases, many of which may share symptoms with schizophrenia and schizoaffective disorder. Genes associated with frontotemporal dementia (26 , 27) , parkinsonism (28 , 29) , and progressive supranuclear palsy (30) and homeobox genes involved in brain development (31) all lie within 17q21. It is interesting that one of the first reports of linkage of a behavioral disorder to this region was for a family in which two of the individuals had been diagnosed during their lifetimes with schizophrenia (30) , and the gene underlying the linkage was later shown to be the microtubule associated protein tau (MAPT) gene, which lies 2 cM from the peak linkage in the current study.
The 17q21 region also contains gene loci for autism, a disease with some overlap in features with schizophrenia. Two genome screens on independent samples, conducted with families from the Autism Genetic Research Exchange, each showed a peak lod score for families with unaffected females at the 17q11-17q21 region (32 , 33) , and in the latter study, additional fine mapping narrowed the peak to the 17q21 region (with a peak lod score of 4.1 less than 1 cM from our peak lod score). These studies used a broad phenotype for autism that included individuals who did not have the classic age at onset of autism.
Finally, it is also worth noting that two other neuropsychiatric phenotypes, bipolar disorder (34) and systemic lupus erythematosus (35) , have demonstrated evidence of linkage to 17q21.
Chromosome 15q22-24
The second region of interest from the current genome screen was at the 15q22-24 region. Like chromosome 17q21, this locus has not had attention called to it in any previous genome screens for schizophrenia to our knowledge, but it was one of the few loci that were suggested as potential loci for schizophrenia in a meta-analysis (21) . Although evidence has been found of linkage between 15q and schizophrenia (36) , the locus was at 15q13-14. A combined analysis using subjects with schizophrenia and bipolar disorder found evidence of linkage to a nearby locus (15q26) (37) . One group has suggested the 15q14-21 region as a chromosomal location showing potential overlap of schizophrenia and autism (38) . Finally, in the mid-1990s, there were two reports suggesting that 15q23-24 was a region of potential significance for studies of schizophrenia and autism (39 , 40) .
Conclusion
This second genome-wide linkage screen for schizophrenia in a Latino population provides two new loci of relevance to schizophrenia genetics in populations of Mexican and Central American ancestry. The locations of these linkage peaks present implications concerning potential overlap of schizophrenia, frontotemporal dementia, and autism. Whether these disorders share specific genes or whether they are showing linkage to different genes within these regions will require association-based analyses of specific candidate genes within these regions (15q22-24 and 17q21). Given the symptomatic overlap between some of the clinical manifestations of these three diseases, as well as studies such as that of Rshetsky et al. (41) (which suggests that “schizophrenia, autism, and bipolar disorder share significant genetic overlaps”), there is certainly ample reason to hypothesize that specific genes in these two regions will have relevance for schizophrenia, autism, frontotemporal dementia, and perhaps even bipolar disorder. In terms of specific genes in this region, given the fact that genes for frontotemporal dementia have been identified in the 17q21 region, these are clear candidates for further study in the Latino population.
The most significant linkage peak in the current study (at 17q) differed from the strongest peak in our first screen of Latino families (1p). Moreover, the results of all three NIMH Human Genetics Initiative studies of sibling pairs (in Latino, U.S. Caucasian and African American, and Han Chinese populations) have identified different linkage peaks as their most prominent loci. Combined analyses using all of these study populations, which were collected by using similar methods and diagnostic procedures, may be of interest in identifying additional loci that did not stand out in any individual study. As a group, this series of studies confirms that stochastic variation in human populations most often leads to different genomic localizations in different samples in studies of a complex disorder such as schizophrenia. Moreover, recent studies also suggest that results from different ethnic populations may yield very different localizations for the genes of relevance to those populations.
Taken as a whole, the results of the different schizophrenia studies from the NIMH Human Genetics Initiative support the notion that schizophrenia is caused by several different genes of small effect and there is no single gene of major effect causing this illness. On the positive side, each of these studies has presented evidence for particular chromosomal locations for genes of relevance to schizophrenia, and they have therefore provided the first clues to identification of genes in these regions. Genome-wide association analyses of schizophrenia are now under way and should aid in identifying gene variants of small effect. These studies may use linkage findings from the current set of studies to help prioritize the chromosomal regions on which they should focus their efforts. For Latino populations of Mexican and Central American descent, the loci of greatest interest must now be considered to be chromosome 1pter-p36 (from our previously published genome screen) and 17q21 (from the current genome screen). Association-based analyses and fine mapping should help to further identify the specific loci in these regions that contribute to schizophrenia in this population.
1. Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, Gunnarsdottir S, Ivarsson O, Chou TT, Hjaltason O, Birgisdottir B, Jonsson H, Gudnadottir VG, Gudmundsdottir E, Bjornsson A, Ingvarsson B, Ingason A, Sigfusson S, Hardardottir H, Harvey RP, Lai D, Zhou M, Brunner D, Mutel V, Gonzalo A, Lemke G, Sainz J, Johannesson G, Andresson T, Gudbjartsson D, Manolescu A, Frigge ML, Gurney ME, Kong A, Gulcher JR, Petursson H, Stefansson K: Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet 2002; 71:877–892Google Scholar
2. Chumakov I, Blumenfeld M, Guerassimenko O, Cavarec L, Palicio M, Abderrahim H, Bougueleret L, Barry C, Tanaka H, La Rosa P, Puech A, Tahri N, Cohen-Akenine A, Delabrosse S, Lissarrague S, Picard F-P, Maurice K, Essioux L, Millasseau P, Grel P, Debailleul V, Simon A-M, Caterina D, Dufaure I, Malekzadeh K, Belova M, Luan J-J, Bouillot M, Sambucy J-L, Primas G, Saumier M, Boubkiri N, Martin-Saumier S, Nasroune M, Peixoto H, Delaye A, Pinchot V, Bastucci M, Guillou S, Chevillon M, Sainz-Fuertes R, Meguenni S, Aurich-Costa J, Cherif D, Gimalac A, Van Duijn C, Gauvreau D, Ouellette G, Fortier I, Raelson J, Sherbatich T, Riazanskaia N, Rogaev E, Raeymaekers P, Aerssens J, Konings F, Luyten W, Macciardi F, Sham PC, Straub RE, Weinberger DR, Cohen N, Cohen D: Genetic and physiological data implicating the new human gene G72 and the gene for D-amino acid oxidase in schizophrenia. Proc Natl Acad Sci USA 2002; 99:13675–13680Google Scholar
3. Straub RE: Jiang Y, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, Cesare AJ, Gibberman A, Wang X, O’Neill A, Walsh D, Kendler KS: Genetic variation in the 6p223 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am J Hum Genet 2002; 71:337–348Google Scholar
4. Brzustowicz LM, Hodgkinson KA, Chow EW, Honer WG, Bassett AS: Location of a major susceptibility locus for familial schizophrenia on chromosome 1q21-q22. Science 2000; 288:678–682Google Scholar
5. Pimm J, McQuillin A, Thirumalai S, Lawrence J, Quested D, Bass N, Lamb G, Moorey H, Datta SR, Kalsi G, Badacsonyi A, Kelly K, Morgan J, Punukollu B, Curtis D, Gurling H: The Epsin 4 gene on chromosome 5q, which encodes the clathrin-associated protein enthoprotin, is involved in the genetic susceptibility to schizophrenia. Am J Hum Genet 2005; 76:902–907Google Scholar
6. Abecasis GR, Burt RA, Hall D, Bochum S, Doheny KF, Lundy SL, Torrington M, Roos JL, Gogos JA, Karayiorgou M: Genomewide scan in families with schizophrenia from the founder population of Afrikaners reveals evidence for linkage and uniparental disomy on chromosome 1. Am J Hum Genet 2004; 74:403–417Google Scholar
7. Sklar P, Pato MT, Kirby A, Petryshen TL, Medeiros H, Carvalho C, Macedo A, Dourado A, Coelho I, Valente J, Soares MJ, Ferreira CP, Lei M, Verner A, Hudson TJ, Morley CP, Kennedy JL, Azevedo MH, Lander E, Daly MJ, Pato CN: Genome-wide scan in Portuguese island families identifies 5q31–5q35 as a susceptibility locus for schizophrenia and psychosis. Mol Psychiatry 2004; 9:213–218Google Scholar
8. Escamilla MA, Ontiveros A, Nicolini H, Raventos H, Mendoza R, Medina R, Munoz R, Levinson D, Peralta JM, Dassori A, Almasy L: A genome-wide scan for schizophrenia psychosis susceptibility loci in families of Mexican and Central American ancestry. Am J Med Genet B Neuropsychiatr Genet 2007; 144:193–199Google Scholar
9. Riley B, Kendler KS: Molecular genetic studies of schizophrenia. Eur J Hum Genet 2006; 14:669–680Google Scholar
10. Nurnberger JI Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T; NIMH Genetics Initiative: Diagnostic Interview for Genetic Studies: rationale, unique features, and training. Arch Gen Psychiatry 1994; 51:849–859Google Scholar
11. Maxwell E: The Family Interview for Genetic Studies Manual. Washington, DC, National Institute of Mental Health, Intramural Research Program, Clinical Neurogenetics Branch, 1992Google Scholar
12. Sobel E, Papp JC, Lange K: Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet 2002; 70:496–508Google Scholar
13. Dyke B: PEDSYS: A Pedigree Data Management System. San Antonio, Tex, Southwest Foundation for Biomedical Research, Population Genetics Laboratory, 1992Google Scholar
14. Abecasis GR, Cherny SS, Cookson WO, Cardon LR: Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 2002; 30:97–101Google Scholar
15. Kong A, Cox NJ: Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 1997; 61:1179–1188Google Scholar
16. Feingold E, Brown PO, Siegmund D: Gaussian models for genetic linkage analysis using complete high-resolution maps of identity by descent. Am J Hum Genet 1993; 53:234–251Google Scholar
17. Lander E, Kruglyak L: Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 1995; 11:241–247Google Scholar
18. Campos-Sánchez R, Barrantes R, Silva S, Escamilla M, Ontiveros A, Nicolini H, Mendoza R, Munoz R, Raventos H: Genetic structure analysis of three Hispanic populations from Costa Rica, Mexico and the Southwest of the United States, using Y-STRs and mtDNA markers. Hum Biology 2006; 78:551–563Google Scholar
19. Risch N, Merikangas K: The future of genetic studies of complex human diseases. Science 1996; 273:1516–1517Google Scholar
20. Brzustowicz LM: Size matters: the unexpected challenge of detecting linkage in large cohorts (editorial). Am J Psychiatry 2007; 164:192–194Google Scholar
21. Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, Williams NM, Schwab SG, Pulver AE, Faraone SV, Brzustowicz LM, Kaufmann CA, Garver DL, Gurling HM, Lindholm E, Coon H, Moises HW, Byerley W, Shaw SH, Mesen A, Sherrington R, O’Neill FA, Walsh D, Kendler KS, Ekelund J, Paunio T, Lönnqvist J, Peltonen L, O’Donovan MC, Owen MJ, Wildenauer DB, Maier W, Nestadt G, Blouin JL, Antonarakis SE, Mowry BJ, Silverman JM, Crowe RR, Cloninger CR, Tsuang MT, Malaspina D, Harkavy-Friedman JM, Svrakic DM, Bassett AS, Holcomb J, Kalsi G, McQuillin A, Brynjolfson J, Sigmundsson T, Petursson H, Jazin E, Zoëga T, Helgason T: Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: schizophrenia. Am J Hum Genet 2003; 73:34–48Google Scholar
22. Moldin SO: NIMH Human Genetics Initiative: 2003 update. Am J Psychiatry 2003; 160:621–622Google Scholar
23. Suarez BK, Duan J, Sanders AR, Hinrichs AL, Jin CH, Hou C, Buccola NG, Hale N, Weilbaecher AN, Nertney DA, Olincy A, Green S, Schaffer AW, Smith CJ, Hannah DE, Rice JP, Cox NJ, Martinez M, Mowry BJ, Amin F, Silverman JM, Black DW, Byerley WF, Crowe RR, Freedman R, Cloninger CR, Levinson DF, Gejman PV: Genomewide linkage scan of 409 European-ancestry and African American families with schizophrenia: suggestive evidence of linkage at 8p233-p212 and 11p131-q141 in the combined sample. Am J Hum Genet 2006; 78:315–333Google Scholar
24. Faraone SV, Hwu H-G, Liu C-M, Chen WJ, Tsuang M-M, Liu S-K, Shieh M-H, Hwang T-J, Ou-Yang W-C, Chen C-Y, Chen C-C, Lin J-J, Chou FH-C, Chueh C-M, Liu W-M, Hall M-H, Su J, Van Eerdewegh P, Tsuang MT: Genome scan of Han Chinese schizophrenia families from Taiwan: confirmation of linkage to 10q22.3. Am J Psychiatry 2006; 163:1760–1766Google Scholar
25. Cardno AG, Holmans PA, Reese MI, Jones LA, McCarthy GM, Hamshere ML, Williams NM, Norton N, Williams HJ, Fenton I, Murphy KC, Sanders RD, Gray MY, O’Donovan MC, McGuffin P, Owen MJ: A genomewide linkage study of age of onset in schizophrenia. Am J Med Genet 2001; 105:439–445Google Scholar
26. Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, Hackett J, Adamson J, Lincoln S, Dickson D, Davies P, Petersen RC, Stevens M, de Graaff E, Wauters E, van Baren J, Hillebrand M, Moosse M, Kwon JM, Nowotny P, Che LK, Norton J, Morris JC, Reed LA, Trojanowski J, Basun H, Lannfelt L, Neystat M, Fahn S, Dark F, Tannenberg T, Dodd PR, Hayward N, Kwok JB, Schofield PR, Andreadis A, Snowden J, Crauford D, Neary D, Owen F, Oostra BA, Hardy J, Goate A, van Swieten J, Mann D, Lynch T, Heutink P: Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 1998; 393:702–705Google Scholar
27. Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S, Cannon A, Dwosh E, Neary D, Melquist S, Richardson A, Dickson D, Berger Z, Eriksen J, Robinson T, Zehr C, Dicke CA, Crook R, McGowan E, Mann D, Boeve B, Feldman H, Hutton M: Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 2006; 442:916–919Google Scholar
28. Mailliot C, Bussiere T, Hamdane M, Sergeant N, Caillet ML, Delacourte A, Buee L: Pathological tau phenotypes: the weight of mutations, polymorphisms, and differential neuronal vulnerabilities. Ann NY Acad Sci 2000; 920:107–114Google Scholar
29. Martin ER, Scott WK, Nance MA, Watts RL, Hubble JP, Koller WC, Lyons K, Pahwa R, Stern MB, Colcher A, Hiner BC, Jankovic J, Ondo WG, Allen FH, Goetz CG, Small GW, Masterman D, Mastaglia F, Laing NG, Stajich JM, Ribble RC, Booze MW, Rogala A, Hauser MA, Zhang F, Gibson RA, Middleton LT, Roses AD, Jaines JL, Scott BL, Pericak-Vance MA, Vance JM: Association of single-nucleotide polymorphisms of the tau gene with late-onset Parkinson disease. JAMA 2001; 286:2245–2250Google Scholar
30. Wilhelmsen KC, Lynch T, Pavlou E, Higgins M, Hygaard TG: Localization of disinhibition-dementia-parkinsonism-amyotrophy complex to 17q21–22. Am J Hum Genet 1994; 55:1159–1165Google Scholar
31. Acampora D, D’Esposito M, Faiella A, Pannese M, Migliaccio E, Morelli F, Stornaiuolo A, Nigro V, Simeone A, Boncinelli E: The human HOX gene family. Nucleic Acids Res 1989; 17:10385–10402Google Scholar
32. Stone JL, Merriman B, Cantor RM, Yonan AL, Gilliam TC, Geschwind DH, Nelson SF: Evidence for sex-specific risk alleles in autism spectrum disorder. Am J Hum Genet 2004; 75:1117–1123Google Scholar
33. Cantor RM, Kono N, Duvall JA, Alvarez-Retuerto A, Stone JL, Alarcon M, Nelson SF, Geschwind DH: Replication of autism linkage: fine-mapping peak at 17q21. Am J Hum Genet 2005; 76:1050–1056Google Scholar
34. Ewald H, Wikman FP, Teruel BM, Buttenschon HN, Torralba M, Als TD, El Daoud TJ, Flint TJ, Jorgensen TH, Blanco L, Kruse TA, Orntoft TF, Mors O: A genome-wide search for risk genes using homozygosity mapping and microarrays with 1,494 single-nucleotide polymorphisms in 22 eastern Cuban families with bipolar disorder. Am J Med Genet B Neuropsychiatr Genet 2005; 133:25–30Google Scholar
35. Cantor RM, Yuan J, Napier S, Kono N, Grossman JM, Hahn BH, Tsao BP: Systemic lupus erythematosus genome scan: support for linkage at 1q23, 2q33, 16q12-13, and 17q21-23 and novel evidence at 3p24, 10q23-24, 13q32, and 18q22-23. Arthritis Rheum 2004; 50:3203–3210Google Scholar
36. Freedman R, Coon H, Myles-Worsley M, Orr-Urtreger A, Olincy A, Davis A, Polymeropoulos M, Holik J, Hopkins J, Hoff M, Rosenthal J, Waldo MC, Reimherr F, Wender P, Yaw J, Young DA, Breese CR, Adams C, Patterson D, Adler LE, Kruglyak L, Leonard S, Byerley W: Linkage of a neurophysiological deficit in schizophrenia to a chromosome 15 locus. Proc Natl Acad Sci USA 1997; 94:587–592Google Scholar
37. Vazza G, Bertolin C, Scudellaro E, Vettori A, Boaretto F, Rampinelli S, De Sanctis G, Perini G, Peruzzi P, Mostacciulo ML: Genome-wide scan supports the existence of a susceptibility locus for schizophrenia and bipolar disorder on chromosome 15q26. Mol Psychiatry 2007; 12:87–93Google Scholar
38. Chagnon YC: Shared susceptibility region on chromosome 15 between autism and catatonia. Int Rev Neurobiol 2006; 72:165–178Google Scholar
39. Goodman AB: A family history study of schizophrenia spectrum disorders suggests new candidate genes in schizophrenia and autism. Psychiatr Q 1994; 65:287–297Google Scholar
40. Goodman AB: Amyotrophic lateral sclerosis, lymphoma and autism in schizophrenia. Schizophr Res 1994; 11:102Google Scholar
41. Rshetsky A, Wajngurt D, Park N, Zheng T: Probing genetic overlap among complex human phenotypes. Proc Natl Acad Sci USA 2007; 104:11694–11699Google Scholar