Modification of Heritability for Educational Attainment and Fluid Intelligence by Socioeconomic Deprivation in the UK Biobank
Abstract
Objective:
Socioeconomic factors have been suggested to influence the effect of education- and intelligence-associated genetic variants. However, results from previous studies on the interaction between socioeconomic status and education or intelligence have been inconsistent. The authors sought to assess these interactions in the UK Biobank cohort of 500,000 participants.
Methods:
The authors assessed the effect of socioeconomic deprivation on education- and intelligence-associated genetic variants by estimating the single-nucleotide polymorphism (SNP) heritability for fluid intelligence, educational attainment, and years of education in subsets of UK Biobank participants with different degrees of social deprivation, using linkage disequilibrium score regression. They also generated polygenic scores with LDpred and tested for interactions with social deprivation.
Results:
SNP heritability increased with socioeconomic deprivation for fluid intelligence, educational attainment, and years of education. Polygenic scores were also found to interact with socioeconomic deprivation, where the effects of the scores increased with increasing deprivation for all traits.
Conclusions:
These results indicate that genetics have a larger influence on educational and cognitive outcomes in more socioeconomically deprived U.K. citizens, which has serious implications for equality of opportunity.
Potential interactions between socioeconomic factors and genetic variants that are related to education and cognitive ability may have serious implications for millions of people in light of the increases in income inequality across many of the industrialized nations over the past 30 years (1). Educational attainment, measured either in years of education or whether an individual has attended college or university, constitutes a complex genetic trait that correlates with cognitive abilities, such as intelligence. It is strongly linked to quality-of-life measurements such as subjective well-being (2), paid employment (2), overall health (3, 4), mortality (5), and obesity (6). A series of large-scale genome-wide association studies (GWASs) (7–12), the most recent on 1.1 million participants (11), have identified more than 1,000 independent educational attainment–associated SNPs and estimated the SNP heritability—that is, the proportion of phenotypic variance explained by all SNPs included in a GWAS—at approximately 22% (12).
Findings from the most recent GWAS indicate the presence of gene-environment interactions (11). Genetic effect sizes were found to differ between the cohorts that were used for meta-analyses, and differences in SNP heritability for educational attainment between cohorts were also reported (11). For example, the SNP heritability was observed to be larger in cohorts from countries where income inequality is larger (11), which suggests that socioeconomic factors related to income inequality influence genetic effects. This has also been studied in twin cohorts, in which both higher (13–16) and lower (17) heritability has been reported in children from less deprived backgrounds, whereas other studies have found no detectable difference in heritability across the socioeconomic gradient (18–20). These findings are limited by relatively small sample sizes, and comparisons between studies are further complicated by differences in sample characteristics, such as participants’ age, year of birth, and the like, as well as methodological differences (see Table S1 in the online supplement).
In this study, we sought to leverage the large sample size of the UK Biobank cohort to assess the interaction between socioeconomic deprivation and intelligence or educational attainment. We used the Townsend deprivation index (TDI) as a proxy for socioeconomic deprivation in UK Biobank participants (Table 1). TDI scores incorporate data on employment, car and home ownership, and household overcrowding and are generated for each U.K. national census output zone (21). UK Biobank participants were assigned TDI scores on their initial assessment according to their residential postal codes and the most recent national census. Educational attainment was assessed in all participants via touchscreen questionnaire. As a measurement of intelligence, we used fluid intelligence assessments, which were available for approximately 37% of all UK Biobank participants.
| Characteristic | Combined Cohort (N=362,498) | 1st Quintile (N=72,829) | 2nd Quintile (N=72,843) | 3rd Quintile (N=72,865) | 4th Quintile (N=72,860) | 5th Quintile (N=72,865) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | N | % | N | % | |
| Female | 195,067 | 53.9 | 38,825 | 53.3 | 39,020 | 53.6 | 39,358 | 54.0 | 39,644 | 54.4 | 37,984 | 52.1 |
| College or university graduate | 113,355 | 31.6 | 25,655 | 35.8 | 23,459 | 32.7 | 22,238 | 31.0 | 22,743 | 31.7 | 19,042 | 26.6 |
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Age (years) | 56.9 | 8.0 | 57.3 | 7.8 | 57.4 | 7.8 | 57.1 | 7.9 | 56.5 | 8.1 | 56.0 | 8.2 |
| Year of birth | 1951 | 8.0 | 1951 | 7.8 | 1951 | 7.8 | 1951 | 8.0 | 1952 | 8.1 | 1952 | 8.2 |
| Years of education | 13.8 | 5.1 | 14.5 | 4.9 | 14.1 | 5.0 | 13.9 | 5.1 | 13.8 | 5.2 | 13.0 | 5.3 |
| Townsend deprivation index | −1.57 | 2.94 | −4.75 | 0.56 | −3.47 | 0.32 | −2.33 | 0.37 | −0.54 | 0.71 | 3.25 | 1.83 |
| Fluid intelligence score | 6.22 | 2.10 | 6.42 | 2.03 | 6.31 | 2.05 | 6.24 | 2.07 | 6.30 | 2.12 | 5.91 | 2.20 |
| N | % | N | % | N | % | N | % | N | % | N | % | |
| Fluid intelligence score questionnaire responders | 131,688 | 36.3 | 23,035 | 31.6 | 26,741 | 36.7 | 26,965 | 37.0 | 28,646 | 39.3 | 26,114 | 35.8 |
| Professional qualifications questionnaire responders | 359,094 | 99.1 | 71,796 | 98.6 | 71,751 | 98.5 | 71,767 | 98.5 | 71,817 | 98.6 | 71,528 | 98.2 |
TABLE 1. Characteristics of the full cohort and Townsend deprivation index quintiles of the UK Biobank cohort
METHODS
UK Biobank Cohort
The UK Biobank is a retrospective and prospective cohort study of 502,492 citizens across the United Kingdom who were recruited between 2006 and 2010. Ethical approval to collect participant data was given by the North West Multicentre Research Ethics Committee, the National Information Governance Board for Health and Social Care, and the Community Health Index Advisory Group. All participants provided signed consent to participate in UK Biobank. Participants were 37–73 years of age at the time of recruitment, and they provided biological samples as well as detailed information via touchscreen questionnaire. To minimize the effect of population stratification, we filtered the cohort for participants who self-identified as British for our analyses. Additional filtering included only participants classified as Caucasian by principal component analysis (data field 22006). Genetic relatedness pairing was provided by the UK Biobank (data field 22011). We excluded related individuals based on kinship data (estimated genetic relationship >0.044), individuals with poor call rate (<95%), individuals with high heterozygosity (data field 22010), and individuals with sex errors (data field 22001). After filtering, data from 362,498 participants remained.
Educational attainment was assessed in the touchscreen questionnaire by the question “Which of the following qualifications do you have?” (data field 6138). Participants could respond “College or university degree,” “A levels/AS levels or equivalent,” “O levels/GCSEs or equivalent,” “CSEs or equivalent,” “NVQ or HND or HNC or equivalent,” “Other professional qualifications, e.g., nursing, teaching,” “None of the above,” and “Prefer not to answer.” Educational attainment was coded as a binary variable (1 for participants who reported having attended college or university and 0 for participants who reported any other level of qualification). Participants who preferred not to answer or who answered “None of the above” were set as missing, resulting in 359,094 participants with informative data for analyses. Years of education was inferred from the same variable, using an application of the International Standard Classification of Education (ISCED-97) on the United Kingdom’s educational qualifications (22). “College or University degree” was coded as 20 years of education, “A levels/AS levels or equivalent” as 13 years, “O levels/GCSEs or equivalent” as 10 years, “CSEs or equivalent” as 10 years, “NVQ or HND or HNC or equivalent” as 19 years, “Other professional qualifications, e.g., nursing, teaching” as 15 years, and “None of the above” as 7 years, resulting in 362,498 participants with informative data on years of education.
Fluid intelligence scores (data field 20016) for each participant were estimated through a battery of 13 questions given via touchscreen questionnaire (see Table S2 in the online supplement). Participants were given 2 minutes to answer as many questions as they were able to. Participants were also able to skip questions. Fluid intelligence scores were calculated as the unweighted sum of the number of correct answers given to the 13 questions, and thus ranged from 0 to 13. Questions that remained unanswered after the allotted 2 minutes were scored as zero. In total, 165,471 participants completed the fluid intelligence test at the assessment centers. After filtering, fluid intelligence scores were available for 131,688 participants. Fluid intelligence was also assessed remotely via a web-based questionnaire (data field 20191), where an additional question was included. A total of 123,651 participants took the web-based test, of whom 46,699 had already performed tests at the assessment centers. We elected to use only the results from the assessment center tests, since these tests were undertaken under more controlled conditions and were administered to the largest number of participants.
Information on the participants’ socioeconomic status was available via the TDI, which is a measure of material deprivation that is based on unemployment rates, non–car ownership, non–home ownership, and household overcrowding. A higher index corresponds to a higher degree of deprivation, which is analogous to lower socioeconomic status. Data for the TDI were collected during the national census and calculated for each census output area. Participants were scored using data from the preceding national census and were assigned a score according to the output area in which their postal code is located. The cohort was stratified into quintiles based on TDI (Table 1).
Genotyping
Genotyping of UK Biobank participants had been performed on two genotyping microarrays, the UK BiLEVE and UK Biobank Axiom genotyping arrays. Approximately 50,000 samples were genotyped on the UK BiLEVE array and approximately 450,000 samples on the Axiom array, which both genotype approximately 850,000 variants. The two arrays are highly similar, with >95% overlap. SNPs had been imputed up to a total of 97,056,775 genetic variants. SNPs were filtered for >0.01% minor allele frequency, no deviation from Hardy-Weinberg equilibrium (p>10−20), and per-variant and per-sample missing genotype rates <5%. After filtering, 34,453,499 SNPs were included for analyses. However, only a subset of these variants was used for subsequent analyses, as described below.
Genome-Wide Association Studies
Association tests for all traits were performed with PLINK, version 1.90b3n (23). Association of SNPs with fluid intelligence scores and years of education was modeled using linear regression. Age, sex, a batch variable for the two genotyping arrays used in the UK Biobank, and 15 principal components were included as covariates. Associations between SNPs and educational attainment were tested with logistic regression modeling, since educational attainment was coded as a binary variable. The same covariates as in the linear regression models were included. The cohort was stratified into quintiles by TDI, and GWASs were performed separately for each trait in each quintile. GWAS was also performed for each trait in the combined cohort, that is, all TDI quintiles together. We also performed a GWAS for TDI, using the same covariates as above, in order to generate summary statistics for genetic correlation.
SNP Heritability and Genetic Correlation
SNP heritabilities and genetic correlations between traits and strata were estimated using LDSC, version 1.0.0 (24). Linkage disequilibrium (LD) scores from 1000 Genomes European data (downloaded from https://data.broadinstitute.org/alkesgroup/LDSCORE/eur_w_ld_chr.tar.bz2) were used to weight regression coefficients for correlated SNPs in the LD score regression analyses. SNP heritabilities were estimated for each trait in each TDI quintile as well as for each trait in the full cohort.
Polygenic Scores
Previous studies on gene-environment interactions have demonstrated that interaction effects of individual SNPs are generally small, which makes them difficult to detect, even with large sample sizes (25). In contrast, polygenic scores provide a good opportunity for interaction analyses, as they are able to aggregate the effects of many SNPs into one variable. Therefore, polygenic scores for educational attainment, years of education, and fluid intelligence were estimated with LDpred, version 0.9.9 (26). First, we filtered for SNPs included in HapMap v3 (N=1,217,311), which are commonly well imputed. The UK Biobank cohort was randomly split into a training (33%) and a testing (67%) set. We used a larger testing set to increase the likelihood of detecting statistically significant differences between quintiles.
We performed a GWAS for each trait in the training set to estimate the effect for each SNP. We then used LDpred to coordinate the GWAS summary statistics with the genotype data for a reference sample of 5,000 randomly selected unrelated Caucasian UK Biobank participants who self-reported as British. In this step, SNPs are filtered for allele frequency (minor allele frequency >1%) and SNPs with ambiguous or nonmatching nucleotides between the GWAS summary statistics and the reference sample are removed. After filtering, 930,923 SNPs remained for fluid intelligence and 1,142,611 SNPs remained for educational attainment and years of education. In a second step, LDpred was used to calculate LD-based weights for the effect of each SNP. This step requires specification of a window around each SNP, given in number of SNPs, as a basis for LD adjustment. As recommended by the authors of the LDpred program (26), we specified an LD window of approximately 1,000 kilobases, which corresponds to an LD radius of M/3000, where M is the number of SNPs used for the analyses. This resulted in an LD radius of 310 for fluid intelligence and 380 for years of education and educational attainment.
LDpred was then used to calculate posterior means of effects that are conditional on the LD pattern of the reference data set, as well as a genetic architecture prior. The prior has two parameters: the heritability explained by the genotypes, which is estimated from GWAS summary statistics, and the fraction of causal markers (rho), that is, the fraction of markers with nonzero effects. The fractions of causal markers are unknown for the traits in this study. As a default, LDpred calculates posterior means based on seven different values for rho: 1.0, 0.3, 0.1, 0.03, 0.01, 0.003, and 0.001. The optimal rho can then be selected according to the performance of the polygenic scores in the training set. For fluid intelligence score and years of education, the most optimal model was determined by comparing how linear models improved when the polygenic score was added. This was performed by calculating the squared semipartial correlation coefficients (ΔR2) for linear models that included each score as well as sex, age, TDI, a batch variable for the two genotyping arrays that were used in UK Biobank, and 15 principal components. For educational attainment, which was coded as a binary trait, the score with the best discriminatory capacity was determined by calculating the area under the receiver operating characteristic curve, using the same covariates and the pROC package (27) in R. Optimal models were achieved at rho=0.03 for all three traits (see Tables S3–S5 in the online supplement). Allelic scoring of the remaining 67% of participants who were not included in the GWAS was then performed with PLINK using the “−score” function and the SNP weights from the LDpred model with rho=0.03.
Gene-Environment Interaction Analyses
Gene-environment interactions were tested with regression modeling in R, version 3.5.1 (28), using the glm function. Linear regression was used for modeling fluid intelligence score and years of education, and logistic regression modeling was used for educational attainment. Educational attainment, fluid intelligence, or years of education was set as the response variable. Polygenic scores, age, sex, TDI, a batch variable, and 15 principal components were included as covariates. Each model also included terms for interactions between all covariates in accordance with recommendations by Keller (29). Three traits were tested, and a p value <0.017 (0.05/3) was considered significant.
We also tested for interactions between TDI and individual SNPs for educational attainment, despite the limited power to detect gene-environment interactions for individual SNPs. We elected to use results for educational attainment for these analyses, since the GWAS on educational attainment in the combined cohort yielded the largest number of genome-wide significant SNPs (N=188; p<5 × 10−8). Lead SNPs were tested for interaction with TDI using PLINK, version 1.90b3n, with the “−interaction” flag, which performs multiple linear regression modeling of models that include SNP-by-covariate interaction terms. β estimates for SNP-by-TDI interaction terms that were generated from these models were tested for deviation from zero using Student’s t test. Bonferroni correction was used to adjust for multiple testing, and p values <2.7 × 10−4 (0.05/188 SNPs) were considered significant.
Sensitivity Analyses: Resampling of TDI Subgroups With Similar Mean Polygenic Scores
A difference in mean polygenic scores was observed between TDI quintiles (see Table S6 in the online supplement). To ensure that such differences would not drive the relative change in estimated heritability between quintiles, we performed a set of sensitivity analyses. We resampled TDI-stratified subsets so that all subsets had a similar polygenic score. First, we defined a density function, fT (t), common to all polygenic scores such that

 : fT (t) > 0}, fXk (t), k = 1,…,n denotes the density function that describes the distribution of the polygenic score in the kthn-quantile of TDI, and where A is a renormalization constant. It is assumed that the support for all density functions is equal, such that SX1 = … = SXn = ST. We stratified the cohort by TDI into three parts (see Table S7 in the online supplement). Three parts were chosen in order to attain similar sample sizes after resampling, as in the primary analyses with five quintiles, and thus conserve statistical power. In practice, the resampling was performed as follows: First, we created three histograms from the polygenic scores of each TDI tertile by dividing the range in polygenic score in a number of bins with bin size 0.01, and counted the number of individuals in each bin. By definition, there is an equal number of individuals in each TDI tertile, so no normalization was needed. Second, we selected the minimum number of individuals in each bin by comparing the three histograms using the pmin() function in R, version 3.5.3. The resulting histogram corresponds to the common density function, defined above. Third, we randomly removed individuals from each of the three original TDI subsets such that the number of remaining individuals in each resampled subset matched the number of individuals in the common histogram. This resampling was performed using the sample() function in R, without replacement, and resulted in three TDI subsets with equal distributions in polygenic score. About 5%−6% of individuals were removed by this procedure, depending on the trait. Finally, GWAS was performed in each subset, and the summary statistics were used to estimate SNP heritability similarly to the primary analyses.
: fT (t) > 0}, fXk (t), k = 1,…,n denotes the density function that describes the distribution of the polygenic score in the kthn-quantile of TDI, and where A is a renormalization constant. It is assumed that the support for all density functions is equal, such that SX1 = … = SXn = ST. We stratified the cohort by TDI into three parts (see Table S7 in the online supplement). Three parts were chosen in order to attain similar sample sizes after resampling, as in the primary analyses with five quintiles, and thus conserve statistical power. In practice, the resampling was performed as follows: First, we created three histograms from the polygenic scores of each TDI tertile by dividing the range in polygenic score in a number of bins with bin size 0.01, and counted the number of individuals in each bin. By definition, there is an equal number of individuals in each TDI tertile, so no normalization was needed. Second, we selected the minimum number of individuals in each bin by comparing the three histograms using the pmin() function in R, version 3.5.3. The resulting histogram corresponds to the common density function, defined above. Third, we randomly removed individuals from each of the three original TDI subsets such that the number of remaining individuals in each resampled subset matched the number of individuals in the common histogram. This resampling was performed using the sample() function in R, without replacement, and resulted in three TDI subsets with equal distributions in polygenic score. About 5%−6% of individuals were removed by this procedure, depending on the trait. Finally, GWAS was performed in each subset, and the summary statistics were used to estimate SNP heritability similarly to the primary analyses.Sample Truncation Bias Due to Stratification on TDI
The observed difference in distribution (mean, variance, etc.) of polygenic scores between TDI quintiles (see Table S6 in the online supplement) may partly be a manifestation of possible sample truncation bias (30). This type of bias occurs when stratification is performed on the outcome or on a variable that is affected by the outcome. In our analyses, we stratified by TDI, which may be affected by the outcome—for example, educational attainment. Sample truncation bias may result in attenuated main effect estimates. If the cohort is stratified on the outcome, this would lead to a truncated error term (30). Hence, the distributions of the model residuals in the different strata should both differ from each other and deviate from the distribution of the unstratified residuals, that is, in the presence of sample truncation bias. In order to test this, we regressed the outcome variables on the polygenic score in each quintile of TDI and in the unstratified sample, with the same covariate terms as in the full models, excluding all main and interaction terms with TDI, and compared the corresponding residuals.
Collider Bias Due to Pleiotropic Effects
Stratification of the UK Biobank cohort on the basis of TDI could potentially result in collider bias (31), since TDI is associated with both the cognitive ability trait and the polygenic score for that trait. TDI may be considered a collider in our model if educational attainment and the polygenic score for educational attainment both independently and directly influence TDI. But this would require that the SNPs in the polygenic score show horizontal pleiotropy, that is, that they are associated with both TDI and educational attainment, but through independent causal pathways (32).
Methods to test for horizontal pleiotropy in an ensemble of SNPs have been developed for Mendelian randomization analyses. We used the HEIDI-outlier approach, which is implemented in the Mendelian randomization package gsmr (33), to identify pleiotropic SNPs. This method requires a set of robustly associated SNPs, rather than all SNPs used to construct the polygenic score from LDpred. We therefore used a previous GWAS on educational attainment in 1.1 million participants (11) to select and extract summary statistics for 1,207 independent SNPs (R2<0.1) that were significantly associated (p<5 × 10−8) with educational attainment. We then estimated the corresponding effects of these SNPs on TDI in the UK Biobank and checked for pleiotropic outliers by running gsmr (33) with the HEIDI-outlier analysis flag set to “true.” Of the 1,207 original SNPs, 61 were found to be horizontally pleiotropic outliers by the HEIDI-outlier analysis (see Figure S1 in the online supplement). We removed those 61 SNPs and generated a polygenic score from the remaining 1,146 nonpleiotropic SNPs. We then estimated both the effects of the collider-free polygenic score on educational attainment in each TDI quintile of the UK Biobank and the interaction effect between the polygenic score and TDI, similarly to the primary analyses. For comparison, similar analyses were performed for a polygenic score generated from all 1,207 SNPs.
Statistical Analysis
The change in SNP heritability with TDI quintiles for the three traits was assessed by simple linear regression: h2 ∼ TDI, assuming homoscedastic errors in heritability. The mean TDI was taken as the representative values for TDI in each quintile. The p value for the trend in heritability was given by the p value for the estimated slope (Student’s t test). A binomial test was used to calculate the probability for at least the observed number of SNPs with a nominally significant interaction term, given the null hypothesis of no interaction.
RESULTS
Genome-wide association analyses were performed for educational attainment, years of education, and fluid intelligence in five UK Biobank subgroups, stratified into quintiles based on TDI (Table 1), as well as in the combined cohort. The summary statistics from each GWAS were used to estimate SNP heritabilities and genetic correlations. Across TDI quintiles, SNP heritabilities ranged from 21.6% to 31.0% for fluid intelligence score, 13.4% to 26.4% for educational attainment, and 13.8% to 24.3% for years of education (Figure 1). SNP heritabilities for all three traits were observed to increase with socioeconomic deprivation (fluid intelligence: p for trend=8.29 × 10−4; educational attainment: p=6.20 × 10−3; years of education: p=1.07 × 10−2) (see Figure 1; see also Table S8 in the online supplement).
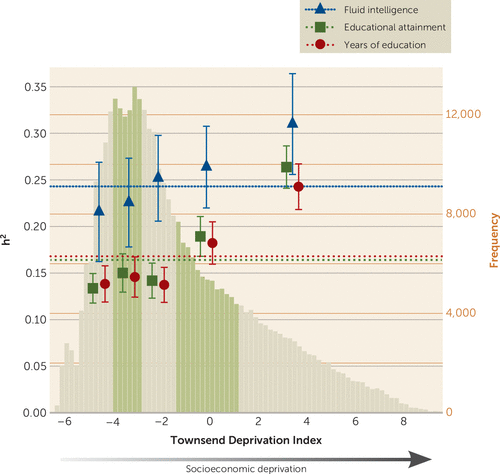
Strong genetic correlations were observed between quintiles within each trait (see Table S9 in the online supplement). Correlations between the fifth quintile and both the first and second quintiles for educational attainment and years of education were below unity (see Table S9), which is consistent with the imperfect genetic correlations and differences in heritability that were reported between cohorts with different levels of income inequality in the recent GWAS on 1.1 million participants (11).
Genetic correlation between educational attainment and years of education was approximately unity (see Table S10 in the online supplement), which is expected, since both variables are derived from the same underlying questionnaire data. Both traits are nonetheless included in our analyses, as they both frequently occur in the literature on education and genetics.
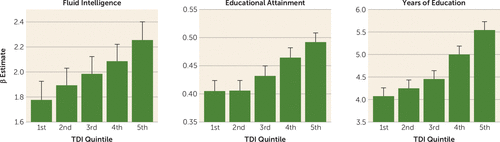
Polygenic scores were observed to interact with TDI for educational attainment (p=1.08 × 10−10), years of education (p=1.9 × 10−12), and fluid intelligence (p=1.10 × 10−4), where the effect of the polygenic scores increased with socioeconomic deprivation for all traits (Figure 2; see also Tables S11–S13 in the online supplement), which is consistent with the observations of increasing SNP heritability with social deprivation.
In addition to investigating the interaction between polygenic scores and TDI, we also tested for the interaction between individual SNPs and TDI for educational attainment. In the combined-cohort GWAS for educational attainment, we identified 188 genome-wide significant lead SNPs that were tested for interaction with TDI. We were unable to observe any significant interaction after Bonferroni correction. However, evidence of interaction at the nominal level was observed for 25 SNPs (see Table S14 in the online supplement), which is a larger number than would be expected by chance (binomial test, p=8.9 × 10−6). These results suggest that there is an underlying pattern of interactions, but that the power to detect individual interactions is limited when SNPs are investigated individually.
Sensitivity Analyses
Small differences in the mean value and distribution of polygenic scores between TDI quintiles was observed (see Table S6 in the online supplement). These differences may potentially introduce bias in our heritability estimates as a result of truncation of the outcome variable caused by the stratification on TDI (30). However, by resampling TDI-stratified subsets of UK Biobank participants with similar mean and distribution of polygenic scores (see Table S7 in the online supplement), we found that the SNP heritabilities were still higher with higher TDI, consistent with the results from the primary analyses (see Figure S2 in the online supplement). We therefore conclude that the observed differences in heritability between quintiles were not caused by differences in polygenic score between quintiles. Moreover, the distributions of the model residuals for all TDI quintiles were found to closely resemble those for the unstratified sample (see Figures S3–S5 in the online supplement), which indicates that all error distributions are similar. Any sample truncation bias (30) due to stratification on TDI should therefore be limited in our analyses.
We were concerned about obtaining biased interaction effects when we stratified by TDI as a result of collider bias. In our analyses, genetic correlation was present between TDI and our phenotypes of interest. The strongest correlation was between TDI and educational attainment (see Table S10 in the online supplement). While a genetic correlation could be due to vertical pleiotropy (SNPs acting through the same causal pathway), correlation due to horizontal pleiotropy (SNPs acting through different pathways) is of primary concern for collider bias. When a subset of 1,207 lead SNPs for educational attainment from a previous GWAS (11) were investigated, only 61 were found to be horizontally pleiotropic outliers (see Figure S1 in the online supplement), which suggests that these SNPs could affect TDI by independent pathways and make TDI a collider when these SNPs are included in the polygenic score. However, the remaining 1,146 SNPs did not show any evidence of horizontal pleiotropy, and the effects among the TDI quintiles of a “collider-free” polygenic score that only included these 1,146 SNPs showed no significant difference compared with a polygenic score that also included the 61 potentially pleiotropic SNPs (see Table S15 in the online supplement). Significant interactions with TDI were observed for both polygenic scores, similar to what was observed for the primary polygenic score from LDpred. This suggests that the interaction effect is not driven by collider bias and that our results are robust and remain valid.
DISCUSSION
In our analyses, the effects of educational attainment- and fluid intelligence–associated genetic variants differ by degree of socioeconomic deprivation. This results in higher heritabilities for these traits in participants from more socially deprived areas of the United Kingdom. The results are consistent with the diathesis-stress model (17), in which low socioeconomic status represents a more challenging environment during childhood and adolescence, which interacts with individuals’ genetic predispositions for educational attainment and cognitive ability through gene-environment interactions. According to this model, a socially deprived individual’s genetic predispositions for educational attainment and fluid intelligence have a larger role on outcomes, in comparison with individuals from a more supportive environment.
The results from this study contrast with those from twin studies that have demonstrated an increase in heritability for cognitive abilities with higher childhood socioeconomic status (13–16, 34) (see Table S1 in the online supplement), which is in the opposite direction of what we observed here. However, several twin studies were unable to replicate this interaction (16–20, 35). A recent meta-analysis that aimed to resolve this discrepancy (36) showed that studies of twins in the United States were almost consistent in being able to demonstrate increasing heritability for intelligence-related traits with childhood socioeconomic factors, such as level of parental education and income. In contrast, studies of western European or Australian populations were consistent in not being able to report any gene-environment interactions, with one exception: a study of Dutch adolescent twins found higher heritability for cognitive ability in children from mothers with lower levels of education (37), which is consistent with the findings of our study. National differences thus appear to play a role in moderating interaction effects between genetic factors and intelligence-related traits. Possible reasons for this discrepancy between the United States and other countries include differences in access to and quality of education across the socioeconomic spectrum, as well as differences in access to welfare support systems and access to and affordability of health care. In addition, pedagogical interventions that are more common in the United States, such as aptitude testing and selection of children for gifted and talented programs and specialized high schools, may also influence the effect of genetic variants and lead to differences in heritability across the socioeconomic spectrum.
Educational attainment has previously been shown also to be influenced by the environment within a family, an effect that is separate from the genetic variants that are inherited. In a recent study, it was even shown that both transmitted (nature) and nontransmitted (nurture) parental genetic alleles affect the educational outcome of offspring via the family environment (38). Whether nurture effects also depend on socioeconomic factors is less clear. However, interactions between nurture and socioeconomic factors could possibly contribute to part of the interaction effects that were observed in our study.
In contrast to previous studies, this is the first to use summary statistics from GWASs from a cross-sectional cohort to study the interaction between socioeconomic factors and education- and intelligence-associated genetic variants. This has enabled a manyfold increase in sample size, which should result in more confident results compared with previous studies. However, our results would benefit from being replicated in an independent cohort. Examining these effects in a large cohort of U.S. participants would also allow for confirmation of possible contrasts between the United States and other countries. However, because of the use of GWAS summary statistics, we restricted our analyses to common SNPs and SNPs that are known to be of high imputation quality. It is possible that rare genetic variation and de novo mutations also interact with socioeconomic factors. Previous studies have shown, for example, that the enrichment of ultra-rare damaging mutations in conserved genes is associated with having fewer years of education (39). However, because of the bias toward common variants on the genotyping arrays in the UK Biobank, we were not able to address the effect of rare variants in our study. Whole genome sequencing is now performed in many studies and will be a valuable resource for further quantification of gene-environment interactions of rare or even family-specific variants.
A limiting factor in this study is the lack of information on UK Biobank participants’ socioeconomic deprivation during childhood and adolescence, which is the most critical period for educational attainment and cognitive development. The TDI scores that we used as a measure of the participants’ socioeconomic situation were based on the most recent national census before participants were interviewed, and their correlation with socioeconomic deprivation during childhood and adolescence may be limited because of the social mobility of UK Biobank participants. A study performed in 1972 (40) showed that approximately 50% of U.K. citizens born between 1938 and 1947 ended up in a different class of employment from their parents. The median year of birth for UK Biobank participants is 1950. However, social mobility has also been observed to differ depending on the social class to which an individual is born (41). A recent combined study of four British birth cohorts, from 1946, 1958, 1970, and 1980–1984 (41), showed that children of parents with professional or managerial positions were more likely to end up in the same social class as their parents, in comparison with children of working-class parents. The observed heterogeneity of heritability between TDI quintiles is therefore likely to partially correlate with socioeconomic deprivation during childhood and adolescence, particularly in low-deprivation segments of society where social mobility has been observed to be lower (41). In addition, a “healthy volunteer” bias has also been documented in the UK Biobank (42), which led to a slightly skewed distribution of phenotypic and demographic traits. For example, UK Biobank participants are generally leaner and less likely to be smokers compared with the general population of the United Kingdom. The rate of participation was also higher among women, older age groups, and persons living in less socioeconomically deprived areas (42). The observations reported in this study are not likely to have emerged as a result of this bias. However, quantitative estimates such as prevalence, incidence, and effect sizes of individual SNPs and interactions may not be directly generalizable to the U.K. population.
CONCLUSIONS
The results from our study imply that socioeconomic deprivation interacts with human biology to magnify the genetic effects on educational and cognitive outcomes. Even though this study was performed in a cohort from the United Kingdom, the results may generalize to other countries with similar socioeconomic gradients and systems of education. Of course, socioeconomic deprivation is associated with an array of factors that may be causal for the interactions observed in this study, such as high stress (43), poor diet (44, 45), higher rates of childhood obesity (46), substance abuse (47), mental illness (48), poor access to health care (49), and lower-quality schools (50). The effects of these factors may also be compounded by national or municipal policies related to funding for welfare and social services. Regardless of which factors are causal, socioeconomic deprivation constitutes a modifiable environmental factor that can be alleviated through social policies.
1.
2. : Education and the subjective quality of life . J Health Soc Behav 1997 ; 38 : 275 – 297 Crossref, Medline, Google Scholar
3. : The links between education and health . Am Sociol Rev 1995 ; 60 : 719 – 745 Crossref, Google Scholar
4. : What does education do to our health? Measuring the effects of education on health and civic engagement. Proceedings of the Copenhagen Symposium 2006 , pp 355–363 (http://www.oecd.org/edu/innovation-education/37425763.pdf) Google Scholar
5. , : Socioeconomic inequalities in health in 22 European countries . N Engl J Med 2008 ; 358 : 2468 – 2481 Crossref, Medline, Google Scholar
6. , : Childhood predictors of adult obesity: a systematic review . Int J Obes Relat Metab Disord 1999 ; 23 ( suppl 8 ): S1 – S107 Medline, Google Scholar
7. , : Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N=112 151) . Mol Psychiatry 2016 ; 21 : 758 – 767 Crossref, Medline, Google Scholar
8. , : Genome-wide association study identifies 74 loci associated with educational attainment . Nature 2016 ; 533 : 539 – 542 Crossref, Medline, Google Scholar
9. , : Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function . Nat Commun 2018 ; 9 : 2098 Crossref, Medline, Google Scholar
10. , : Genome-wide association meta-analysis of 78,308 individuals identifies new loci and genes influencing human intelligence . Nat Genet 2017 ; 49 : 1107 – 1112 Crossref, Medline, Google Scholar
11. , : Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals . Nat Genet 2018 ; 50 : 1112 – 1121 Crossref, Medline, Google Scholar
12. , : GWAS of 126,559 individuals identifies genetic variants associated with educational attainment . Science 2013 ; 340 : 1467 – 1471 Crossref, Medline, Google Scholar
13. : Genetic and environmental influences on vocabulary IQ: parental education level as moderator . Child Dev 1999 ; 70 : 1151 – 1162 Crossref, Medline, Google Scholar
14. , : Socioeconomic status modifies heritability of IQ in young children . Psychol Sci 2003 ; 14 : 623 – 628 Crossref, Medline, Google Scholar
15. , : Emergence of a gene × socioeconomic status interaction on infant mental ability between 10 months and 2 years . Psychol Sci 2011 ; 22 : 125 – 133 Crossref, Medline, Google Scholar
16. : Childhood socioeconomic status amplifies genetic effects on adult intelligence . Psychol Sci 2013 ; 24 : 2111 – 2116 Crossref, Medline, Google Scholar
17. : Environmental moderators of genetic influence on verbal and nonverbal abilities in early childhood . Intelligence 2005 ; 33 : 643 – 661 Crossref, Google Scholar
18. , : Gene-environment interaction in adults’ IQ scores: measures of past and present environment . Behav Genet 2008 ; 38 : 348 – 360 Crossref, Medline, Google Scholar
19. , : Does parental education have a moderating effect on the genetic and environmental influences of general cognitive ability in early adulthood? Behav Genet 2010 ; 40 : 438 – 446 Crossref, Medline, Google Scholar
20. , : Socioeconomic status (SES) and children’s intelligence (IQ): in a UK-representative sample SES moderates the environmental, not genetic, effect on IQ . PLoS One 2012 ; 7 : e30320 https://doi.org/ Crossref, Medline, Google Scholar
21. : Health and Deprivation: Inequality and the North. London , Croom Helm , 1988 Google Scholar
22. :
23. : PLINK: an R package for linking mixed-format tests using IRT-based methods . J Stat Softw 2010 ; 35 : 1 – 33 Crossref, Medline, Google Scholar
24. , : LD score regression distinguishes confounding from polygenicity in genome-wide association studies . Nat Genet 2015 ; 47 : 291 – 295 Crossref, Medline, Google Scholar
25. , : Gene-environment interaction study for BMI reveals interactions between genetic factors and physical activity, alcohol consumption, and socioeconomic status . PLoS Genet 2017 ; 13 : e1006977 https://doi.org/ Crossref, Medline, Google Scholar
26. , : Modeling linkage disequilibrium increases accuracy of polygenic risk scores . Am J Hum Genet 2015 ; 97 : 576 – 592 Crossref, Medline, Google Scholar
27. , : pROC: an open-source package for R and S+ to analyze and compare ROC curves . BMC Bioinformatics 2011 ; 12 : 77 https://doi.org/ Crossref, Medline, Google Scholar
28.
29. : Gene × environment interaction studies have not properly controlled for potential confounders: the problem and the (simple) solution . Biol Psychiatry 2014 ; 75 : 18 – 24 Crossref, Medline, Google Scholar
30. : Social experimentation, truncated distributions, and efficient estimation . Econometrica 1977 ; 45 : 919 Crossref, Google Scholar
31. , : A robust example of collider bias in a genetic association study . Am J Hum Genet 2016 ; 98 : 392 – 393 Crossref, Medline, Google Scholar
32. : Evaluating the potential role of pleiotropy in Mendelian randomization studies . Hum Mol Genet 2018 ; 27 ( R2 ): R195 – R208 Crossref, Medline, Google Scholar
33. , : Causal associations between risk factors and common diseases inferred from GWAS summary data . Nat Commun 2018 ; 9 : 224 https://doi.org/ Crossref, Medline, Google Scholar
34. : Genotype by environment interaction in adolescents’ cognitive aptitude . Behav Genet 2007 ; 37 : 273 – 283 Crossref, Medline, Google Scholar
35. , : Heritability of word recognition in middle-aged men varies as a function of parental education . Behav Genet 2005 ; 35 : 417 – 433 Crossref, Medline, Google Scholar
36. : Large cross-national differences in gene × socioeconomic status interaction on intelligence . Psychol Sci 2016 ; 27 : 138 – 149 Crossref, Medline, Google Scholar
37. : Breastfeeding, maternal education, and cognitive function: a prospective study in twins . Behav Genet 2009 ; 39 : 616 – 622 Crossref, Medline, Google Scholar
38. , : The nature of nurture: effects of parental genotypes . Science 2018 ; 359 : 424 – 428 Crossref, Medline, Google Scholar
39. , : Ultra-rare disruptive and damaging mutations influence educational attainment in the general population . Nat Neurosci 2016 ; 19 : 1563 – 1565 Crossref, Medline, Google Scholar
40. : Social class mobility in modern Britain: changing structure, constant process . Journal of the British Academy 2016 ; 4 : 89 – 111 Crossref, Google Scholar
41. , : The mobility problem in Britain: new findings from the analysis of birth cohort data . Br J Sociol 2015 ; 66 : 93 – 117 Crossref, Medline, Google Scholar
42. , : Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population . Am J Epidemiol 2017 ; 186 : 1026 – 1034 Crossref, Medline, Google Scholar
43. : Childhood poverty, chronic stress, self-regulation, and coping . Child Dev Perspect 2013 ; 7 : 43 – 48 Crossref, Google Scholar
44. , : Socio-economic differences in diet, physical activity, and leisure-time screen use among Scottish children in 2006 and 2010: are we closing the gap? Public Health Nutr 2017 ; 20 : 951 – 958 Crossref, Medline, Google Scholar
45. , : Trends in socio-economic inequalities in the Scottish diet: 2001–2009 2015 ; 18 : 2970 – 2980 Google Scholar
46. : Childhood obesity and overweight prevalence trends in England: evidence for growing socioeconomic disparities . Int J Obes 2010 ; 34 : 41 – 47 Crossref, Google Scholar
47. , : Associations of social and material deprivation with tobacco, alcohol, and psychotropic drug use, and gender: a population-based study . Int J Health Geogr 2007 ; 6 : 50 https://doi.org/ Crossref, Medline, Google Scholar
48. , : Socioeconomic status and adolescent mental disorders . Am J Public Health 2012 ; 102 : 1742 – 1750 Crossref, Medline, Google Scholar
49. , : Measures of social deprivation that predict health care access and need within a rational area of primary care service delivery . Health Serv Res 2013 ; 48 : 539 – 559 Crossref, Medline, Google Scholar
50. : Schools in disadvantaged areas: recognising context and raising quality (CASE Paper 076). London, London School of Economics, Centre for Analysis of Social Exclusion, 2004 Google Scholar



