Down-Regulation of SIRT1 Gene Expression in Major Depressive Disorder
Tothe Editor: Major depressive disorder is one of the most prevalent mental health disorders and a leading cause of disability worldwide. Although major depressive disorder has a relatively high heritability, which has been estimated to be around 0.32 (1), the genes contributing to risk of major depressive disorder remain largely unknown. A recent large-scale genome-wide association study (including 5,303 case subjects and 5,337 control subjects) showed that genetic variants near the SIRT1 gene are significantly associated with major depressive disorder (2), suggesting that SIRT1 may play a role in the pathogenesis of major depressive disorder. To test if the dysregulation of SIRT1 has a role in major depressive disorder, we measured the expression level of SIRT1 in subjects with major depressive disorder and in comparison subjects.
Fifty unrelated major depressive disorder subjects who were not taking medication and 50 comparison subjects (age and sex-matched) were recruited from Shanghai Mental Health Center, and peripheral blood samples were collected between 7:00 a.m. and 9:00 a.m. from fasting patients and healthy comparison subjects. Detailed information about sample collection, diagnosis, RNA extraction, reverse transcription, real-time quantitative polymerase chain reaction (PCR), and statistical analyses can be found in the data supplement accompanying the online edition of this letter. Glyceraldehyde-3-phosphate dehydrogenase was used as an internal control, and the 2-ΔΔCt method (3) was used to analyze the fold change of quantitative PCR data.
We found that SIRT1 expression is significantly down-regulated in the peripheral blood of patients with major depressive disorder compared with comparison subjects (decreased by 37%) (p=3.98×10−8) (Figure 1). To further validate our results, we examined SIRT1 expression in the peripheral blood of major depressive disorder cases and control subjects using data from a recent large-scale major depressive disorder expression study (4). Again, we found that SIRT1 expression is significantly down-regulated in remitted major depressive disorder cases (cases, N=635; control subjects, N=331; p=5.50×10−3) compared with controls. These consistent results suggest that dysregulation of SIRT1 expression may have a role in major depressive disorder. Interestingly, a recent animal model study also found that dysregulation of SIRT1 signaling has a critical role in depression-like behaviors (5). These convergent lines of evidence support the idea that dysregulation of SIRT1 expression may play an important role in major depressive disorder and suggest that SIRT1 may be a potential target for future therapeutics and diagnostics. Although these results suggest an important role for SIRT1 in major depressive disorder, it should be noted that major depressive disorder is most likely a polygenic phenotype and that many other loci remain to be discovered beyond SIRT1.
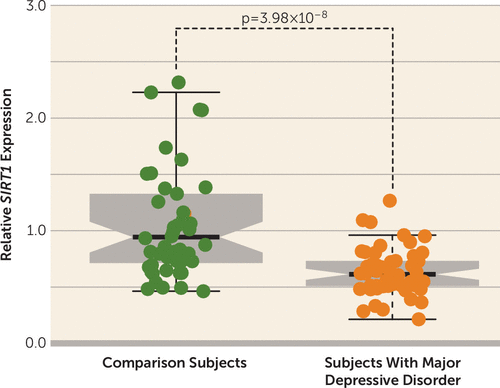
FIGURE 1. Significant Down-Regulation of SIRT1 Expression in Major Depressive Disordera
a Expression of SIRT1 is decreased by 37% in cases of major depressive disorder relative to comparison subjects.
1 : Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry 2000; 157:1552–1562Link, Google Scholar
2
3 : Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25:402–408Crossref, Medline, Google Scholar
4 : Gene expression in major depressive disorder. Mol Psychiatry 2016; 21:339–347Crossref, Medline, Google Scholar
5 : Hippocampal sirtuin 1 signaling mediates depression-like behavior. Biol Psychiatry (Epub ahead of print, Jan 30, 2016)Google Scholar



