Localization of Bipolar Susceptibility Locus by Molecular Genetic Analysis of the Chromosome 12q23-q24 Region in Two Pedigrees With Bipolar Disorder and Darier’s Disease
Abstract
OBJECTIVE: The authors previously reported two families (pedigrees 324 and 5501) in which Darier’s disease—a rare, autosomal dominant skin disease—and bipolar disorder cosegregate. In each of these families there is complete cosegregation of mood disorder with a segment of chromosome 12q23-q24, consistent with the existence of a highly penetrant dominant variant. Here molecular genetic analyses aimed at localizing and identifying the susceptibility gene in this region are reported. METHOD: In the two families, the authors undertook 1) linkage and haplotype studies using 45 highly polymorphic molecular genetic markers in order to delineate the region of interest and 2) direct analysis of genes within this region. RESULTS: Linkage and haplotype information from the most severely affected individuals defined a region of interest that spanned two neighboring regions of 19 megabases (Mb) (D12S362–D12S1646) and 7 Mb (D12S1718–D12S837). Information from all individuals refined the region of interest to 6.5 Mb (D12S127–D12S1646). Systematic study of the coding and flanking intronic regions of 25 known genes within this latter region failed to identify any highly penetrant autosomal dominant disease-conferring mutations in these pedigrees. CONCLUSIONS: This linkage and haplotype analysis, together with data from several other linkage studies, provides compelling evidence for the existence in the 12q23-q24 region of one or more genes involved in the pathogenesis of bipolar disorder. Further molecular genetic analysis of this region is required to identify the gene(s).
There is convincing evidence from family, twin, and adoption studies for the importance of genes in determining susceptibility to bipolar disorder (1–3). The relative recurrence risk (compared with population risk) for narrowly defined illness (DSM-IV bipolar I disorder) in siblings of a proband, λS, is approximately 8. The monozygotic probandwise concordance rate is approximately 60% (λMZ=60), compared with a dizygotic recurrence risk that is similar to that for siblings (λDZ=8) (4, 5). Within the families of bipolar probands there is a higher than average rate of unipolar depressive disorders (6), indicating that bipolar susceptibility genes can be expressed in a broad spectrum of mood phenotypes. To explain the inheritance of bipolar disorder within the general population, it is necessary to invoke interacting (epistatic) oligo-genes or more complex genetic models (7). However, there have been relatively rare descriptions of families in which inheritance is consistent with the segregation of alleles of major effect (e.g., references 8–11). We have previously described two such families in which there is cosegregation of Darier’s disease (a rare, autosomal dominant skin disease) and major affective disorder: pedigree 324 (12) and pedigree 5501 (13). The Darier’s disease gene was mapped to the 12q23-q24.1 region (14, 15). The cosegregation in pedigrees 324 and 5501 is consistent with genetic linkage between the Darier’s disease gene and a gene that, in these families, confers risk for major mood disorders according to a highly penetrant, autosomal dominant mode of inheritance. The presence of a susceptibility gene for mood disorders in the 12q23-q24 region is supported by linkage studies of bipolar disorder (11, 16–21) and of recurrent major depression (22, 23), an observation consistent with the hypothesis that at least some susceptibility genes contribute to both disorders. As is often the case for complex genetic disorders, the linkage signals span a region of several tens of centimorgans.
The locus causing Darier’s disease has been identified as ATP2A2, which encodes a sarcoplasmic/endoplasmic reticulum calcium-ATPase, SERCA2 (24). One hypothesis that deserves consideration is that in these two pedigrees bipolar disorder could be due to pleiotropic effects of mutations in ATP2A2 (12). This possibility finds some support from our observation of three individuals with both Darier’s disease and bipolar-spectrum mood disorder who had mutations in the same domain of ATP2A2 (25). However, against this are 1) a lack of evidence demonstrating a population-level association between bipolar disorder and Darier’s disease, 2) incomplete cosegregation of bipolar disorder and Darier’s disease in pedigree 5501 (13), demonstrating that the Darier mutation in this family is not on its own sufficient to cause bipolar disorder, and 3) our failure to identify ATP2A2 mutations in bipolar individuals from families showing greater allele sharing at this chromosomal region (26). Thus, the current data suggest that the observed cosegregation is the result of linkage rather than pleiotropy. This is the hypothesis underpinning the current study, in which we 1) delineated the 12q23-q24 region most likely to harbor a bipolar susceptibility gene and 2) systematically studied 25 known, annotated genes within the narrowest region of interest in order to search for mutations that confer risk of bipolar disorder according to a highly penetrant autosomal dominant model in these families.
Method
All subjects in these studies were Caucasians of European origin. All provided written informed consent to participate. Ethical approval was obtained from relevant ethical review committees.
Descriptions of pedigrees 324 and 5501 follow. Other than co-occurrence of Darier’s disease, no unusual clinical characteristics of mood disorders have been experienced by the members of these pedigrees.
Pedigree 324
All members were interviewed by using the Schedule for Affective Disorders and Schizophrenia—Lifetime Version (27), and case notes were obtained. Lifetime best-estimate psychiatric diagnoses were made without knowledge of dermatological diagnoses. A detailed description of this pedigree has been given previously (12). During follow-up one individual was identified as having mild (subclinical) features of Darier’s disease (28)—about which the individual had never been aware—and subsequently developed severe affective illness meeting the DSM-IV criteria for major depressive disorder. The pedigree structure and psychiatric diagnoses are shown in Figure 1.
Pedigree 5501
A modified version of the Schedules for Clinical Assessment in Neuropsychiatry (29) was used to assess the members of this family, and two independent clinicians made best-estimate lifetime consensus diagnoses according to DSM-IV. The pedigree has been described in detail elsewhere (13); the pedigree structure and psychiatric diagnoses are shown in Figure 2.
Haplotype Analysis
Selection of markers
Microsatellite markers across the region were drawn from publicly accessible databases. Primers were designed by using Primer 3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Details of these markers are available on request from the authors. Genetic distances were taken from the Marshfield map (http://research.marshfieldclinic.org/genetics/Map_Markers/maps/IndexMapFrames.html), and physical distances were taken from the Golden Path (November 2002 freeze, http://genome.ucsc.edu/cgi-bin/hgGateway).
Genotyping
DNA was extracted from whole blood according to routine procedures. Microsatellite markers were analyzed on an ABI 377 sequencer (Applied Biosystems, Foster City, Calif.) with GeneScan version 3.1 (Applied Biosystems).
Haplotype and linkage analysis
SIMWALK version 2.83 (30) as implemented at the U.K. Medical Research Council Human Genome Mapping Project Computing Centre (http://www.rfcgr. mrc.ac.uk/Registered/Menu/link-gen.html) was used for data cleaning and maximum likelihood estimation of microsatellite haplotypes unconstrained by phenotypic data. Multiple program runs were performed to confirm the stability and robustness of haplotype assignment. The patterns of haplotype transmission were then examined to determine the haplotypes shared by affected individuals and not shared by unaffected individuals within each family. This was undertaken under each of two phenotypic models.
| 1. | Individuals with mood disorders were designated “affected,” and those without mood disorders were “unaffected.” This defined a “narrowest region of interest.” | ||||
| 2. | Individuals with bipolar I or bipolar II disorder were considered “affected,” but all others were treated as “unknown.” As this classification uses information from only the individuals with the most narrowly defined affection, we believe that this model defines a more conservative “full region of interest.” | ||||
Multipoint parametric linkage analysis was performed by using GENEHUNTER 2.0 (31) under an autosomal dominant model according to each of the two phenotype models. The disease allele frequency was set at 1%, and the phenocopy rate was set at 10%.
Selection of genes for screening
A transcript map across the region we identified as the “narrowest region of interest” was generated by using Ensembl (http://www.ensembl.org/) and the Golden Path (http://genome.ucsc.edu/cgi-bin/hgGateway) and by applying the coding exons prediction package NIX (http://www.hgmp.mrc.ac.uk/Registered/Webapp/nix/) to PAC/BAC sequences located within the same region. The priority for screening the known and predicted genes was decided according to the level of certainty and knowledge about the predicted gene. Here we report an analysis of 25 sequences for which there is unambiguous experimental evidence confirming their status as human genes.
Mutation detection methods
We undertook mutation detection using one core affected individual (with bipolar I disorder) from each pedigree and an unaffected comparison family member. The primers were designed to screen all exons and at least 40 base pairs (bp) of flanking introns by aligning the cDNA sequences from Genbank (http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html) with the corresponding genomic BAC/PAC clones.
Polymerase chain reaction (PCR) was carried out under standard conditions. Sequence variations were detected by using a denaturing high-performance liquid chromatography (DHPLC) system and a DNASep column (Transgenomic, San Jose, Calif.) as described previously (32). Optimal temperatures for the analysis were selected by using DHPLCMelt (http://insertion.stanford. edu/melt.html), with an extra run at 2°C above the highest recommended temperature (32). Samples showing heteroduplexes were sequenced by using dichlororhodamine dye terminator chemistry on an ABI 377 sequencer (Applied Biosystems, Foster City, Calif.). Single nucleotide polymorphisms were identified by comparison of electropherograms and/or by using the PolyPhred computer program (33).
For sequence variants identified in an affected pedigree member, evidence for cosegregation (under the “narrowest region of interest” model) within that family was sought. If there was no cosegregation, the investigation was not taken further. Where cosegregation was seen, a set of unaffected, unrelated white U.K. blood donors of European origin were genotyped by an appropriate PCR-based assay to determine whether the minor allele frequency was compatible with the hypothesis of a rare, disease-conferring mutation specific to one or both of pedigrees 324 and 5501.
Results
Haplotype and Linkage Analyses
A total of 45 microsatellite markers spanning a 75-centimorgan (cM), or 47-megabase (Mb), region from D12S81 to D12S1638 were typed. Figure 3 shows the haplotypes in the key individuals that delineate the regions of interest. Figure 4 shows the linkage results.
Pedigree 324
In investigating the narrowest region of interest it was found that a haplotype spanning 11 cM (9.7 Mb) was shared by all individuals with mood disorders and none of those who were unaffected. The haplotype was bounded by D12S1597 proximally and D12S369 distally.
For the full region of interest, the “affected” haplotype extended across the complete region examined.
Pedigree 5501
In regard to the narrowest region of interest, a haplotype spanning 5 cM (6.5 Mb) was shared by all individuals with mood disorders and none of those who were unaffected. The haplotype was bounded by D12S1127 proximally and D12S1646 distally.
For the full region of interest, the “affected” haplotype in the individuals with bipolar I and II disorders was broader, falling into two regions: D12S362 to D12S1646 (19 Mb) and D12S1718 to D12S837 (7 Mb).
Both the narrow and full regions of interest in pedigree 5501 lay within the respective regions in pedigree 324. It can be seen from Figure 3 and Figure 4 that the haplotype results were consistent with the linkage results.
Gene Screening
We screened a total of 93,740 bp of genomic DNA, of which 42,022 bp was coding sequence and 51,718 bp corresponded to adjacent noncoding regions. We identified 48 variants (Table 1). We also identified a 4-bp deletion within noncoding sequence in OAS3 and a variable-number tandem decameric repeat in the gene P-selectin glycoprotein ligand 1, or SELPG, previously described by Afshar-Kharghan et al. (34). All of the sequence variants detected and described here either did not segregate with disease within these pedigrees or were shown to be common polymorphisms.
Discussion
We undertook molecular genetic studies of two pedigrees in which Darier’s disease and major affective disorder cosegregate, in order to delineate the region of chromosome 12q23-q24 most likely to contain a susceptibility gene for bipolar spectrum illness. In each pedigree the pattern of inheritance of mood disorder was consistent with a major gene model. This is certainly not the usual pattern of inheritance of bipolar disorder (35), and therefore it is important to clarify explicitly the assumptions that underpin the approach we took. We assumed that in each pedigree, a variant exists that acts as a highly penetrant autosomal dominant mutation and that the pathogenic variant(s) (which might be different in the two pedigrees) is rare because otherwise such pedigrees would be common in the population.
If we assume that all individuals with any mood disorder in each family carry the susceptibility variant and that those without such a diagnosis do not (in formal genetic terms, this is a completely penetrant autosomal dominant model without phenocopies), then our linkage and haplotype analyses are consistent in identifying a narrowest region of interest of 5 cM (6.5 Mb) (Figure 3 and Figure 4). Alternatively, if we focus on the most clear-cut bipolar cases (i.e., core bipolar phenotype) and exclude unaffected subjects from the analyses to allow for reduced penetrance, our analyses are consistent in identifying a broader “full region of interest” that spans 36 cM (26 Mb) (Figure 3 and Figure 4). The narrowest region of interest can be considered as our most likely location for harboring the susceptibility variant operating in these pedigrees and, therefore, was the priority segment in which we focused the intensive direct gene analysis. The full region defines the boundaries outside of which it is very unlikely that a susceptibility variant of major effect operates in these pedigrees.
As in all genetic studies, our power to refine the location of susceptibility loci is limited by the number of subjects (i.e., the number of informative meioses within our two pedigrees). Improved localization could be achieved if other, preferably large, families demonstrating linkage in this region can be identified and studied with similar approaches.
As mentioned in the introduction, several other groups have reported evidence supporting linkage of bipolar disorder susceptibility to chromosome 12q23-q24. Which of these findings are compatible with our regions of interest, and what is added by our data? Figure 5 shows schematically our findings and the other positive reports across this region. It is well known that in complex diseases the localization of susceptibility genes is poor, with signals often being tens of centimorgans away from the true locus and regions of positive linkage being broad (36). On this basis, the localization provided by haplotype analysis in our pedigrees consolidates the linkage findings reported by Dawson et al. (16), Ekholm et al. (20), Morissette and colleagues (11), Ewald et al. (9), Curtis et al. (18), and Maziade et al. (21), which all yield evidence either inside or within a few centimorgans of our region. Further, the recent genome scan meta-analysis for bipolar disorder that used the ranking method identified the 12q23 region (i.e., the proximal part of our region of interest) as one of 23 “bins” (out of a total of 120) that achieved nominal statistical significance under at least one model (37). Indeed, our data together with these other findings strongly suggest the presence of at least one susceptibility gene for bipolar disorder within the 26-Mb full region of interest. However, the telomeric localization by linkage disequilibrium reported by Degn et al. (19) lies outside our core phenotype region, raising the possibility that an additional mood disorder susceptibility gene resides within the distal 12q24 region. This possibility may represent the human counterpart of the observation in animal studies that multiple quantitative trait loci may be responsible for linkage peaks in genome scans (e.g., reference 38). It is of particular interest that the linkage signals in the recent genome scans for unipolar depression (22, 23) overlap with, and lie at the centromeric end of, our full region of interest. This suggests that genetic variation in this region may influence susceptibility to both bipolar and unipolar forms of major affective disorder. Our own pedigrees are fully consistent with this possibility in that both include unipolar as well as bipolar cases.
We focused our direct gene analysis on the narrowest region of interest. Within the context of our hypotheses and methods, we can exclude the sequences we examined from harboring a major susceptibility mutation in the two pedigrees examined. There are a number of caveats with regard to these results. First, we examined only the most well-characterized genes in the narrowest region of interest. These data will inform the search by other research groups, but it is important to recognize that additional sequences in the region remain to be studied. Second, we need to consider the adequacy of our mutation detection method. The genes were screened by denaturing high-performance liquid chromatography, which has been proven to be a sensitive method of mutation detection with nearly 100% detection efficiency when used with both the recommended DNA strand melting temperature (RTm) and RTm+2°C (32, 39). We can, thus, be confident that we are unlikely to have failed to detect heterozygous variants within the DNA sequence investigated. Third, we did not systematically examine regions that may regulate gene expression. Our rationale was that with the existing state of annotation of the genome, it is impossible to systematically identify all important regulatory elements, and at any rate, mutations in exons dominate the mutation profile of alleles of major effect. Fourth, these genes have been excluded only under our initial model hypothesizing a highly penetrant dominant mode of inheritance. It is possible that there is a more complex mode of inheritance contributing to bipolar disorder within these pedigrees, and indeed, that is the rationale for using a more relaxed genetic model to define the broader, full region of interest. Exclusion of variants under such a model requires a different strategy involving systematic examination of large groups of cases and comparison subjects to seek either linkage disequilibrium, in the case of common disease variants, or multiple rare disease-associated variants. Therefore, while we are continuing to investigate predicted and partially identified genes within the narrowest region of interest according to the highly penetrant dominant model, as a complementary strategy we are undertaking systematic linkage disequilibrium analysis in outbred study groups, combined with direct analysis of specific candidate genes of high prior interest.
 |
Received July 31, 2003; revision received Sept. 12, 2003; accepted Feb. 9, 2004. From the Department of Psychological Medicine, University of Wales College of Medicine; and the Department of Psychiatry, University of Birmingham, Queen Elizabeth Psychiatric Hospital, Birmingham, U.K. Drs. Green, Elvidge, and Jacobsen contributed equally to the work reported in this article. Address correspondence and reprint requests to Professor Craddock, Neuropsychiatric Genetics Unit, Department of Psychological Medicine, University of Wales College of Medicine, Heath Park, Cardiff CF14 4XN, Wales, U.K.; [email protected] (e-mail). Supported by the Wellcome Trust and the U.K. Medical Research Council. The authors thank all the family members who participated in this study.
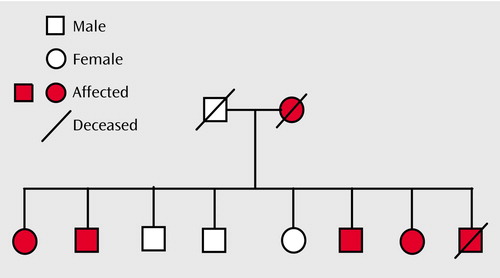
Figure 1. Mood Disorders in a Family With Cosegregating Mood Disorders and Darier’s Disease (Pedigree 324)a
aLifetime DSM-IV diagnoses of mood disorders: bipolar I disorder (N=1), recurrent major depressive disorder (N=4), single episode of major depressive disorder (N=1).
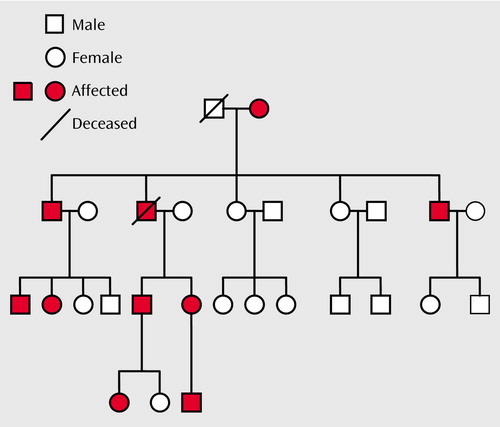
Figure 2. Mood Disorders in a Family With Cosegregating Mood Disorders and Darier’s Disease (Pedigree 5501)a
aLifetime DSM-IV diagnoses of mood disorders: bipolar I disorder (N=3), bipolar II disorder (N=2), bipolar disorder not otherwise specified (N=1), recurrent major depressive disorder (N=1), single episode of major depressive disorder (N=1), depressive disorder not otherwise specified (N=1), adjustment disorder with depressed mood (N=1).
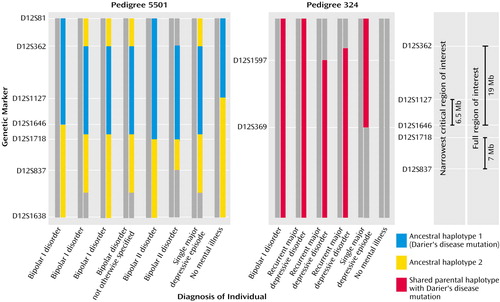
Figure 3. Haplotypes of Two Families With Cosegregating Mood Disorders and Darier’s Disease (Pedigrees 5501 and 324) Based on Microsatellite Markers on Chromosome 12q23-q24.1a
aThe colored boxes indicate the extent of the common haplotype in each pedigree, and the gray boxes indicate all other haplotypes. In pedigree 5501, two ancestral haplotypes were introduced by individual 1.2 (see Figure 2), indicated by the blue boxes (carrying the Darier’s disease mutation) and the yellow boxes. For pedigree 324, the red boxes indicate the shared parental haplotype on which the Darier’s disease mutation lies.
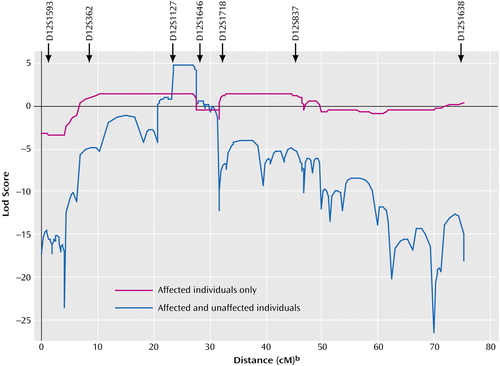
Figure 4. Linkage Analysis Across Chromosome 12q for Two Families With Cosegregating Mood Disorders and Darier’s Disease (Pedigrees 324 and 5501)aa
Multipoint parametric linkage analysis used microsatellite markers from D12S1593 to D12S1638 as described in the text. The blue curve shows results under the phenotype model that uses information from all affected and unaffected individuals. The maximum lod score is 4.77 across the interval D12S1127 to D12S1646 (which includes ATP2A2), identified as the narrowest critical region in our haplotype studies (see text and Figure 3). The purple curve shows the results under the phenotype model that uses information only from those affected by the core bipolar phenotype. The maximum lod score is 1.47 across two intervals that are consistent with the full region of interest identified by the haplotype analysis (see text and Figure 3).
bWith the location of D12S1593 set at zero.
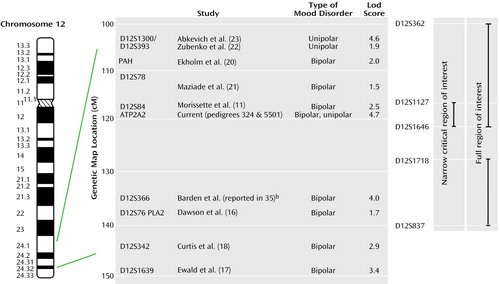
Figure 5. Schematic Representation of Genetic Linkage Findings in Mood Disorder Across a Region of Interest on Chromosome 12qa
aThe genetic map locations in centimorgans are shown according to the Marshfield map (http://research.marshfieldclinic.org/genetics/Map_Markers/maps/IndexMapFrames.html), and relevant genetic markers and gene locations are indicated. The first author of each relevant publication is shown, together with the maximum lod score in support of linkage. “Bipolar” and “unipolar” indicate whether the finding is in families segregating mainly for bipolar or unipolar depression.
bDetails of this result have not been published, and the finding was reported in a chromosome workshop report.
1. Craddock N, Jones I: Genetics of bipolar disorder. J Med Genet 1994; 36:585–594Google Scholar
2. Potash JB, DePaulo JR Jr: Searching high and low: a review of the genetics of bipolar disorder. Bipolar Disord 2000; 2:8–26Crossref, Medline, Google Scholar
3. Baron M: Manic-depression genes and the new millennium: poised for discovery. Mol Psychiatry 2002; 7:342–358Crossref, Medline, Google Scholar
4. Bertelsen A, Harvald B, Hauge M: A Danish twin study of manic-depressive illness. Br J Psychiatry 1977; 130:330–351Crossref, Medline, Google Scholar
5. McGuffin P, Rijsdijk F, Andrew M, Sham P, Katz R, Cardno A: The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch Gen Psychiatry 2003; 60:497–502Crossref, Medline, Google Scholar
6. McGuffin P, Katz R: The genetics of depression and manic-depressive disorder. Br J Psychiatry 1989; 155:294–304Crossref, Medline, Google Scholar
7. Craddock N, Van Eerdewegh P, Reich T: Mathematical limits of multilocus models: the genetic transmission of bipolar disorder. Am J Med Genet 1995; 57:690–702Google Scholar
8. Blackwood DH, He L, Morris SW, McLean A, Whitton C, Thomson M, Walker MT, Woodburn K, Sharp CM, Wright AF, Shibasaki Y, St Clair DM, Porteous DJ, Muir WJ: A locus for bipolar affective disorder on chromosome 4p. Nat Genet 1996; 12:427–430Crossref, Medline, Google Scholar
9. Ewald H, Flint T, Kruse TA, Mors O: A genome-wide scan shows significant linkage between bipolar disorder and chromosome 12q24.3 and suggestive linkage to chromosomes 1p22–21, 4p16, 6q14–22, 10q26 and 16p13.3. Mol Psychiatry 2002; 7:734–744Crossref, Medline, Google Scholar
10. Freimer NB, Reus VI, Escamilla MA, McInnes LA, Spesny M, Leon P, Service SK, Smith LB, Silva S, Rojas E, Gallegos A, Meza L, Fournier E, Baharloo S, Blankenship K, Tyler DJ, Batki S, Vinogradov S, Weissenbach J, Barondes SH, Sandkuijl LA: Genetic mapping using haplotype, association and linkage methods suggests a locus for severe bipolar disorder (BPI) at 18q22-q23. Nat Genet 1996; 12:436–441Crossref, Medline, Google Scholar
11. Morissette J, Villeneuve A, Bordeleau L, Rochette D, Laberge C, Gagne B, Laprise C, Bouchard G, Plante M, Gobeil L, Shink E, Weissenbach J, Barden N: Genome-wide search for linkage of bipolar affective disorders in a very large pedigree derived from a homogeneous population in Quebec points to a locus of major effect on chromosome 12q23–24. Am J Med Genet Neuropsychiatr Genet 1999; 88:567–587Crossref, Medline, Google Scholar
12. Craddock N, Owen M, Burge S, Kurian B, Thomas P, McGuffin P: Familial cosegregation of major affective disorder and Darier’s disease (keratosis follicularis). Br J Psychiatry 1994; 164:355–358Crossref, Medline, Google Scholar
13. Jones I, Jacobsen N, Green EK, Elvidge GP, Owen MJ, Craddock N: Evidence for familial cosegregation of major affective disorder and genetic markers flanking the gene for Darier’s disease. Mol Psychiatry 2002; 7:424–427Crossref, Medline, Google Scholar
14. Craddock N, Dawson E, Burge S, Parfitt L, Mant B, Roberts O, Daniels J, Gill M, McGuffin P, Powell J, Owen M: The gene for Darier’s disease maps to chromosome 12q23-q24.1. Hum Mol Genet 1993; 2:1941–1943Crossref, Medline, Google Scholar
15. Bashir R, Munro CS, Mason S, Stephenson A, Rees JL, Strachan T: Localisation of a gene for Darier’s disease. Hum Mol Genet 1993; 2:1937–1939Crossref, Medline, Google Scholar
16. Dawson E, Parfitt E, Roberts Q, Daniels J, Lim L, Sham P, Nothen M, Propping P, Lanczik M, Maier W, Reuner U, Weissenbach J, Gill M, Powell J, McGuffin P, Owen M, Craddock N: Linkage studies of bipolar disorder in the region of the Darier’s disease gene on chromosome 12q23–24.1. Am J Med Genet 1995; 60:94–102Crossref, Medline, Google Scholar
17. Ewald H, Degn B, Mors O, Kruse TA: Significant linkage between bipolar affective disorder and chromosome 12q24. Psychiatr Genet 1998; 8:131–140Crossref, Medline, Google Scholar
18. Curtis D, Kalsi G, Brynjolfsson J, McInnis M, O’Neill J, Smyth C, Moloney E, Murphy P, McQuillin A, Petursson H, Gurling H: Genome scan of pedigrees multiply affected with bipolar disorder provides further support for the presence of a susceptibility locus on chromosome 12q23-q24, and suggests the presence of additional loci on 1p and 1q. Psychiatr Genet 2003; 13:77–84Medline, Google Scholar
19. Degn B, Lundorf MD, Wang A, Vang M, Mors O, Kruse TA, Ewald H: Further evidence for a bipolar risk gene on chromosome 12q24 suggested by investigation of haplotype sharing and allelic association in patients from the Faroe Islands. Mol Psychiatry 2001; 6:450–455Crossref, Medline, Google Scholar
20. Ekholm JM, Kieseppa T, Hiekkalinna T, Partonen T, Paunio T, Perola M, Ekelund J, Lonnqvist J, Pekkarinen-Ijas P, Peltonen L: Evidence of susceptibility loci on 4q32 and 16p12 for bipolar disorder. Hum Mol Genet 2003; 12:1907–1915Crossref, Medline, Google Scholar
21. Maziade M, Roy MA, Rouillard E, Bissonnette L, Fournier JP, Roy A, Garneau Y, Montgrain N, Potvin A, Cliche D, Dion C, Wallot H, Fournier A, Nicole L, Lavallee JC, Merette C: A search for specific and common susceptibility loci for schizophrenia and bipolar disorder: a linkage study in 13 target chromosomes. Mol Psychiatry 2001; 6:684–693Crossref, Medline, Google Scholar
22. Zubenko GS, Maher B, Hughes HB 3rd, Zubenko WN, Stiffler JS, Kaplan BB, Marazita ML: Genome-wide linkage survey for genetic loci that influence the development of depressive disorders in families with recurrent, early-onset, major depression. Am J Med Genet 2003; 123B(1):1–18Google Scholar
23. Abkevich V, Camp NJ, Hensel CH, Neff CD, Russell DL, Hughes DC, Plenk AM, Lowry MR, Richards RL, Carter C, Frech GC, Stone S, Rowe K, Chau CA, Cortado K, Hunt A, Luce K, O’Neil G, Poarch J, Potter J, Poulsen GH, Saxton H, Bernat-Sestak M, Thompson V, Gutin A, Skolnick MH, Shattuck D, Cannon-Albright L: Predisposition locus for major depression at chromosome 12q22–12q23.2. Am J Hum Genet 2003; 73:1271–1281Crossref, Medline, Google Scholar
24. Sakuntabhai A, Ruiz-Perez V, Carter S, Jacobsen N, Burge S, Monk S, Smith M, Munro CS, O’Donovan M, Craddock N, Kucherlapati R, Rees JL, Owen M, Lathrop M, Monaco AP, Strachan T, Hovnanian A: Mutations in ATP2A2, encoding a Ca2+ pump, cause Darier disease. Nat Genet 1999; 21:271–277Crossref, Medline, Google Scholar
25. Jacobsen NJO, Lyons I, Hoogendoorn B, Burge S, Kwok P, O’Donovan MC, Craddock N, Owen MJ: ATP2A2 mutations in Darier’s disease and their relationship to neuropsychiatric phenotypes. Hum Mol Genet 1999; 8:1631–1636Crossref, Medline, Google Scholar
26. Jacobsen NJ, Franks EK, Elvidge G, Jones I, McCandless F, O’Donovan MC, Owen MJ, Craddock N: Exclusion of the Darier’s disease gene, ATP2A2, as a common susceptibility gene for bipolar disorder. Mol Psychiatry 2001; 6:92–97Crossref, Medline, Google Scholar
27. Endicott J, Spitzer RL: A diagnostic interview: the Schedule for Affective Disorders and Schizophrenia. Arch Gen Psychiatry 1978; 35:837–844Crossref, Medline, Google Scholar
28. Craddock N, McGuffin P, Owen M: Darier’s disease cosegregating with affective disorder (letter). Br J Psychiatry 1994; 165:272Crossref, Medline, Google Scholar
29. Wing JK, Babor T, Brugha T, Burke J, Cooper JE, Giel R, Jablenski A, Regier D, Sartorius N: SCAN: Schedules for Clinical Assessment in Neuropsychiatry. Arch Gen Psychiatry 1990; 47:589–593Crossref, Medline, Google Scholar
30. Sobel E, Lange K: Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 1996; 58:1323–1337Medline, Google Scholar
31. Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES: Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 1996; 58:1347–1363Medline, Google Scholar
32. Jones A, Austin JA, Hansen N, Oefner PJ, Hoogendoorn B, Cheadle JP, O’Donovan MC: Automated optimal temperature selection for denaturing high performance liquid chromatography. Clin Chem 1999; 45:1133–1140Medline, Google Scholar
33. Nickerson DA, Tobe VO, Taylor SL: PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res 1997; 25:2745–2751Crossref, Medline, Google Scholar
34. Afshar-Kharghan V, Diz-Kucukkaya R, Ludwig EH, Marian AJ, Lopez JA: Human polymorphism of P-selectin glycoprotein ligand 1 attributable to variable number of tandem decameric repeats in the mucinlike region. Blood 2001; 97:3306–3307Crossref, Medline, Google Scholar
35. Detera-Wadleigh SD, Barden N, Craddock N, Ewald H, Foroud T, Kelsoe J, McQuillin A: Chromosomes 12 and 16 workshop. Am J Med Genet Neuropsychiatr Genet 1999; 88:255–259Crossref, Medline, Google Scholar
36. Roberts SB, MacLean CJ, Neale MC, Eaves LJ, Kendler KS: Replication of linkage studies of complex traits: an examination of variation in location estimates. Am J Hum Genet 1999; 65:876–884Crossref, Medline, Google Scholar
37. Segurado R, Detera-Wadleigh SD, Levinson DF, Lewis CM, Gill M, Nurnberger JI Jr, Craddock N, DePaulo JR, Baron M, Gershon ES, Ekholm J, Cichon S, Turecki G, et al: Genome scan meta-analysis of schizophrenia and bipolar disorder, part III: bipolar disorder. Am J Hum Genet 2003; 73:49–62Crossref, Medline, Google Scholar
38. Talbot CJ, Radcliffe RA, Fullerton J, Hitzemann R, Wehner JM, Flint J: Fine scale mapping of a genetic locus for conditioned fear. Mamm Genome 2003; 14:223–230Crossref, Medline, Google Scholar
39. Oefner PJ, Underhill PA: DNA Mutation Detection Using Denaturing High-Performance Liquid Chromatography (DHPLC): Current Protocols in Human Genetics Supplement 19. New York, Wiley & Sons, 1998, pp 7.10.1–7.10.12Google Scholar



