GWAS of Suicide Attempt in Psychiatric Disorders and Association With Major Depression Polygenic Risk Scores
Abstract
Objective:
More than 90% of people who attempt suicide have a psychiatric diagnosis; however, twin and family studies suggest that the genetic etiology of suicide attempt is partially distinct from that of the psychiatric disorders themselves. The authors present the largest genome-wide association study (GWAS) on suicide attempt, using cohorts of individuals with major depressive disorder, bipolar disorder, and schizophrenia from the Psychiatric Genomics Consortium.
Methods:
The samples comprised 1,622 suicide attempters and 8,786 nonattempters with major depressive disorder; 3,264 attempters and 5,500 nonattempters with bipolar disorder; and 1,683 attempters and 2,946 nonattempters with schizophrenia. A GWAS on suicide attempt was performed by comparing attempters to nonattempters with each disorder, followed by a meta-analysis across disorders. Polygenic risk scoring was used to investigate the genetic relationship between suicide attempt and the psychiatric disorders.
Results:
Three genome-wide significant loci for suicide attempt were found: one associated with suicide attempt in major depressive disorder, one associated with suicide attempt in bipolar disorder, and one in the meta-analysis of suicide attempt in mood disorders. These associations were not replicated in independent mood disorder cohorts from the UK Biobank and iPSYCH. No significant associations were found in the meta-analysis of all three disorders. Polygenic risk scores for major depression were significantly associated with suicide attempt in major depressive disorder (R2=0.25%), bipolar disorder (R2=0.24%), and schizophrenia (R2=0.40%).
Conclusions:
This study provides new information on genetic associations and demonstrates that genetic liability for major depression increases risk for suicide attempt across psychiatric disorders. Further collaborative efforts to increase sample size may help to robustly identify genetic associations and provide biological insights into the etiology of suicide attempt.
Suicide is a worldwide public health problem accounting for more than 800,000 deaths each year (1). It is the second leading cause of death among young adults, and rates of suicide are far exceeded by suicide attempts, which occur up to 20 times more frequently (1). These statistics represent a huge personal, social, and economic burden, with the Centers for Disease Control and Prevention reporting that suicide costs the U.S. economy $51 billion annually in health care and work-loss related costs (2). These figures highlight the urgent need for improved prevention and treatment; however, progress has been hampered by the lack of reliable methods for predicting suicidality and poor understanding of its biological etiology.
More than 90% of suicide attempters or individuals who die by suicide have a psychiatric disorder, particularly mood disorders, schizophrenia, and substance use disorders (3, 4). Heritability estimates of suicidal behavior from twin studies range from 30% to 55%, and twin and family studies suggest that the genetic etiology of suicide attempt is partially distinct from that of the psychiatric disorders themselves (5, 6). Several genome-wide association studies (GWASs) have been conducted on suicide attempt. These studies have examined individuals with depression or bipolar disorder, comparing attempters with nonattempters and testing for genetic variants that might contribute independently to suicide attempt (7–10). These studies have failed to identify any replicable genetic associations, likely because of limited sample sizes that were underpowered to detect the genetic effects typical for a single-nucleotide polymorphism (SNP). Other GWASs have examined individuals recruited specifically on the basis of suicide attempt or suicide attempters or nonattempters from population-based cohorts, but to date no loci have been definitively implicated (11–14).
Genetic studies have indicated that suicide attempt has a polygenic architecture. Polygenic risk scores for suicide attempt have shown modest predictive ability in independent samples, and small but significant SNP heritability estimates for suicide attempt have been reported (10, 13, 14). These findings are consistent with the presence of small genetic effects that the original GWASs were underpowered to detect at genome-wide significance. In this study, we present the largest GWAS on suicide attempt to date to our knowledge, comparing a total of 6,569 suicide attempters and 17,232 nonattempters with major depressive disorder, bipolar disorder, or schizophrenia from the Psychiatric Genomics Consortium.
Methods
Study Subjects and Phenotype
Study subjects were drawn from 16 cohorts of individuals with major depressive disorder, 21 cohorts of individuals with bipolar disorder, and nine cohorts of individuals with schizophrenia from the Psychiatric Genomics Consortium (PGC), where information on suicide attempt had been collected. Only individuals affected by psychiatric disorders were included, and all psychiatric disorders were defined using structured psychiatric interviews according to international consensus criteria (DSM-IV, ICD-9, or ICD-10) (15–17). Tables S1–S3 in the online supplement summarize the source and inclusion and exclusion criteria for each cohort. All individuals were of European ancestry. Items from structured clinical interviews provided information on self-harm, suicidal ideation and plans, and suicide attempt (see Table S4 in the online supplement). Lifetime suicide attempt was defined across cohorts as a deliberate act of self-harm with at least some intent to result in death. Individuals who did not endorse suicide attempt were included in the nonattempter group, and individuals for whom information on suicide attempt was missing were excluded. Across the data sets for major depressive disorder, bipolar disorder and schizophrenia there were a total of 6,569 suicide attempters and 17,232 nonattempters (Table 1). All study subjects gave written informed consent to participate in the source studies.
| Suicide Attempters | Nonattempters | Female Suicide Attempters | Female Nonattempters | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample | Cohorts (N) | N | % | N | % | N | % | N | % |
| Discovery | |||||||||
| PGCa major depressive disorder | 16 | 1,622 | 16 | 8,786 | 84 | 1,155 | 71 | 5,808 | 66 |
| PGC bipolar disorder | 21 | 3,264 | 37 | 5,500 | 63 | 2,097 | 66 | 2,971 | 56 |
| PGC schizophrenia | 9 | 1,683 | 36 | 2,946 | 64 | 660 | 39 | 924 | 31 |
| Total | 46 | 6,569 | 28 | 17,232 | 72 | 3,912 | 60 | 9,703 | 56 |
| Replication | |||||||||
| UK Biobank (mood disorders)b | 1 | 2,149 | 6 | 35,912 | 94 | 1,521 | 71 | 24,039 | 67 |
| iPSYCH (mood disorders)b | 1 | 4,943 | 24 | 15,849 | 76 | 3,694 | 75 | 10,276 | 65 |
TABLE 1. Sample characteristics of the discovery and replication cohorts
Genotyping, Quality Control, and Imputation
Cohorts were genotyped following their local protocols, after which standardized quality control and imputation were performed centrally using the PGC Rapid Imputation and Computational Pipeline for GWAS (RICOPILI) procedures (https://sites.google.com/a/broadinstitute.org/ricopili/) for each cohort separately. These procedures have been described previously (18–20). Briefly, the quality control parameters for retaining SNPs and study subjects were as follows: missing data for SNP <0.05 (before sample removal), missing data for study subjects <0.02, autosomal heterozygosity deviation of Fhet<0.2, missing data for SNP after sample removal <0.02, difference in missing data for SNP between psychiatric case subjects and healthy control subjects <0.02, SNP Hardy-Weinberg equilibrium of p>10−10 in psychiatric case subjects, and minor allele frequency ≥0.01. Genotype imputation was performed using the prephasing/imputation stepwise approach implemented in IMPUTE2/SHAPEIT (chunk size of 3 Mb and default parameters) to the 1000 Genomes Project reference panel (21–23). SNPs with an imputation INFO score <0.6 in the PLINK genome analysis toolset were excluded. The numbers of SNPs analyzed were 8,482,392, 8,807,005, and 8,814,543 in the major depressive disorder, bipolar disorder, and schizophrenia data sets, respectively.
Statistical Analysis
We performed a GWAS on suicide attempt using PLINK, version 1.9, to compare imputed marker dosages between suicide attempters and nonattempters in each cohort separately using an additive logistic regression model (24). The first five principal components, generated using EIGENSTRAT, were used as covariates in each analysis to control for population stratification (25). No evidence of stratification artifacts or uncontrolled test statistic inflation were observed in the results from any cohort (e.g., λGC was 0.87–1.01). We performed a meta-analysis to obtain GWAS results for suicide attempt in each of the three psychiatric disorders (major depressive disorder, bipolar disorder, and schizophrenia) separately using an inverse variance-weighted fixed-effects model in METAL (26). To increase the sample size, a fixed-effects meta-analysis on suicide attempt was conducted across all three disorders. To investigate depressive suicide attempts, a meta-analysis across major depressive disorder and bipolar disorder was performed. A meta-analysis across bipolar disorder and schizophrenia was used to investigate psychotic suicide attempts.
We used polygenic risk scoring to dissect the genetic relationship between suicide attempt and major depressive disorder, bipolar disorder, and schizophrenia and to test for overlap in the genetic etiology of suicide attempt in each of these three disorders. Table S5 in the online supplement summarizes the polygenic scoring analyses conducted, showing the discovery and test data sets used to investigate these hypotheses. First, we used polygenic risk scores (PRSs) for bipolar disorder, major depression, and schizophrenia to investigate whether individuals who had attempted suicide, and those who had not, differed in genetic liability for the psychiatric disorder that affected them. To ensure no overlap between the discovery and test data sets, we generated PRSs for psychiatric disorders using PGC cohorts not included in the suicide attempt analyses (18–20). The discovery GWAS for bipolar disorder consisted of 8,711 bipolar disorder case subjects and 15,283 control subjects, and the one for schizophrenia included 25,756 schizophrenia case subjects and 35,686 control subjects. The discovery GWAS for major depression was from a recent meta-analysis of PGC major depressive disorder cohorts and samples from deCODE genetics, the Genetic Epidemiology Research on Adult Health and Aging study, iPSYCH, Generation Scotland, and the UK Biobank (18). The phenotype we analyzed included clinically defined cases of major depressive disorder as well as self-reported major depressive disorder symptoms or treatment and thus was referred to as “major depression.” This GWAS had approximately 59,000 case subjects and 112,000 control subjects. We tested PRSs for bipolar disorder, major depression, and schizophrenia for association with suicide attempt compared with nonattempt in the same disorder. Second, based on the results of these analyses, we also tested PRSs for major depression for association with suicide attempt in bipolar disorder and schizophrenia. Third, to investigate genetic overlap in suicide attempt across psychiatric disorders, we used the results of the three GWASs on suicide attempt in major depressive disorder, bipolar disorder, and schizophrenia in turn as discovery studies and tested the PRSs for suicide attempt for association with suicide attempt in the other two disorders.
We used PRSice to generate PRSs, according to standard protocol (27). Briefly, we pruned the GWAS results from each discovery study for linkage disequilibrium using the p-value-informed clumping method, and we selected subsets of SNPs from the results at nine increasingly liberal p value thresholds. We summed sets of alleles, weighted by their log odds ratios from the discovery GWAS, into PRSs for each individual in the test data sets. We investigated PRSs for association with suicide attempt in the test data set using a logistic regression model including five principal components and a covariate for each cohort in the test data set. The amount of variance explained by the PRS (R2) is presented on the liability scale, which accounts for the proportion of cases in the test data set (28).
The Bonferroni-corrected significance threshold for the polygenic scoring analyses was 0.006, adjusted for eight independent tests (see Table S5 in the online supplement). Full details of the PRS calculations, discovery, and test data sets are available in the online supplement.
We assessed the variance in suicide attempt explained by common SNPs (SNP heritability, h2SNP) using genomic-relatedness-based restricted maximum-likelihood (GREML) implemented in GCTA (29). We converted the SNP probabilities to best-guess data with a genotype call probability cutoff of 0.8. We used HapMap 3 SNPs with an INFO score ≥0.6 to calculate the genetic relatedness matrix (GRM) using PLINK 1.9, including individuals with relatedness <0.05 (24). We calculated ancestry-informative principal components using GCTA (29). The GRM was based on a total of 1,166,347 SNPs in the major depressive disorder set, 1,172,705 SNPs in the bipolar disorder data set, and 1,143,070 SNPs in the schizophrenia data set. Covariates included 20 principal components calculated using GCTA (because GRM-based analyses are more sensitive to population stratification than polygenic scoring analyses) and a covariate for each cohort within a disorder. We calculated the h2SNP of suicide attempt in each psychiatric disorder using GCTA (29).
Power Calculations
We used the Genetic Power Calculator to determine the power to detect associations at genome-wide significance (p<5×10−8) for the meta-analysis of 6,569 suicide attempters and 17,232 nonattempters across the three psychiatric disorders (30). This analysis had 78% power to detect an allele with a frequency of 0.2 and an effect size of 1.1 at genome-wide significance. From the GCTA-GREML power calculator, the power to detect an h2SNP of 20% (approximately half the twin heritability estimate) for suicide attempt was 81%, 92%, and 43% in the major depressive disorder, bipolar disorder, and schizophrenia data sets, respectively (31). The statistical power of polygenic risk scoring was calculated using AVENGEME (32, 33). The power of the PRS for the psychiatric disorders to predict suicide attempt in the same disorder was 51% in bipolar disorder, 93% in major depressive disorder, and 78% in schizophrenia, given the h2SNP for these psychiatric disorders calculated from the summary statistics (20%, 10%, and 25%, respectively) and hypothesizing a genetic correlation of 0.5 between the psychiatric disorder and suicide attempt. Given an h2SNP of 20% for suicide attempt, the power of PRSs for suicide attempt to detect a significant difference between attempters and nonattempters in the test data sets ranged from 32% to 64%. The significance threshold for all polygenic risk scoring power calculations was 0.006.
Replication
We tested for replication of genome-wide significant associations with suicide attempt in two independent mood disorder cohorts from the UK Biobank and iPSYCH. The UK Biobank is a population-based prospective study of 501,726 individuals recruited at 23 centers across the United Kingdom (34). Extensive phenotypic data are available for UK Biobank participants from health records and questionnaires, including an online follow-up questionnaire focusing on mental health. Participants were classified as having a mood disorder if they either self-reported a professional diagnosis of depression or bipolar disorder as part of the Mental Health Questionnaire (UK Biobank field 20544) or met criteria for depression or bipolar disorder on Mental Health Questionnaire questions derived from the Composite International Diagnostic Interview. Suicide attempters with mood disorders (N=2,149; 91% with depression) were defined as those who answered yes to the question “Have you ever harmed yourself with the intention to end your life?” (UK Biobank field 20483). Nonattempters with mood disorders were defined as those who reported no self-harm on the Mental Health Questionnaire (N=35,912). Genetic associations with suicide attempt were tested by comparing suicide attempters with mood disorders to nonattempters with mood disorders using BGenie, version 1.2 (35), covarying for six principal components and factors capturing site of recruitment and genotyping batch. Details of the genetic quality control, imputation, and mood disorder criteria are available in the online supplement.
In the iPSYCH cohort, individuals with mood disorders were identified on the basis of ICD-10 codes (F30–F39) from the Danish Psychiatric Central Research Register and the National Registry of Patients, both complete as of December 31, 2016 (36). Suicide attempters with mood disorders (N=4,943, 94% with major depressive disorder) were defined as those with diagnoses of suicide attempt (ICD-10 codes X60–X84, equivalent to intentional self-harm), those with suicide attempt indicated as “reason for contact” and with a main diagnosis of poisoning (ICD-10 codes T39, T42, T43, and T58), or those with a diagnosis in the ICD-10 F chapter as a main diagnosis and report of poisoning by drugs or other substances (ICD-10 codes T36–T50 and T52–T60) or injuries to hand, wrist, or forearm (ICD-10 codes S51, S55, S59, S61, S65, and S69). Individuals who died by suicide according to the Cause of Death Register were also included in the suicide attempt group. Nonattempters were defined as individuals with mood disorders who did not meet any of these criteria (N=15,849). Genetic associations with suicide attempt were tested by comparing suicide attempters with mood disorders to nonattempters with mood disorders, including 10 principal components and genotyping batch as covariates. Quality control for the iPSYCH cohort is described in the online supplement.
Results
Sample Characteristics
The proportion of psychiatric disorder case subjects reporting suicide attempt ranged from 16% in major depressive disorder to 36%−37% in bipolar disorder and schizophrenia (Table 1). For each disorder, there was a higher proportion of females among the suicide attempters than among the nonattempters. Table 1 shows the number and proportion of suicide attempters and nonattempters within each psychiatric disorder. The numbers in the individual PGC cohorts are shown in Tables S6–S8 in the online supplement.
Genome-Wide Association Studies
We performed a GWAS of suicide attempters compared with nonattempters in the major depressive disorder, bipolar disorder, and schizophrenia data sets separately. In the analysis of suicide attempt in major depressive disorder, one SNP reached genome-wide significance: rs45593736 (p=2.61×10−8, odds ratio for the A allele=2.38) (Table 2). This SNP is in an intron of the ARL5B (ADP-ribosylation factor-like 5B) gene, and the A allele has a frequency of 0.02. In the GWAS of suicide attempt in bipolar disorder, an insertion-deletion polymorphism on chromosome 4 met genome-wide significance: chr4_23273116_D (p=1.15×10−8, odds ratio for the deletion=1.29) (Table 2). This is an intronic variant in the noncoding RNA LOC105374524. In the analysis of suicide attempt in schizophrenia, no SNPs reached genome-wide significance, but this analysis had the smallest total sample size.
| Discovery Data Set | Variant | CHR | BP | Allele Tested | Allele Frequency | INFO Score | p | Odds Ratio | 95% CI | Direction in Each Cohortb | Genes (Distance From SNP in kb) | p, UK Biobank Replication Cohortc | p, iPSYCH Replication Cohortc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MDD | rs45593736 | 10 | 18954937 | A | 0.02 | 0.89 | 2.61E–08 | 2.38 | 1.75–3.23 | ?++++–++?+–?++++ | ARL5B (intronic) | 0.985 | 0.776 |
| BD | chr4_23273116_Dd | 4 | 23273116 | D | 0.20 | 0.91 | 1.15E–08 | 1.29 | 1.18–1.41 | ++++++++++++++–+++++ | LOC105374524 | ||
| Mood disorders | rs138689899 | 2 | 128288162 | T | 0.02 | 0.91 | 2.50E–08 | 1.75 | 1.44–2.14 | ++ | IWS1(3.7), MYO7B (107.1) | 0.677 | 0.228 |
| Mood disorders | rs28591567d | 4 | 23253912 | G | 0.22 | 0.95 | 3.11E–08 | 1.19 | 1.12–1.27 | ++ | LOC105374524 | 0.663 | 0.521 |
TABLE 2. Genome-wide significant loci for suicide attempt showing the most significant variant from each genomic regiona
A meta-analysis of the GWAS results for suicide attempt across all three disorders produced no genome-wide significant results (see Figure S4 in the online supplement). In a meta-analysis of suicide attempt in mood disorders (major depressive disorder and bipolar disorder), we found 10 genome-wide significant SNPs from two independent genomic regions (Table 2, Figure 1). The most significant association was rs138689899 on chromosome 2 (p=2.50×10−8, odds ratio for the T allele=1.75). This is an intergenic SNP that lies between the IWS1 and MYO7B genes. The other significant locus was on chromosome 4 in LOC105374524, as found in the bipolar disorder suicide attempt analysis. The most significant SNP was rs28591567 (p=3.11×10−8, odds ratio for the G allele=1.19, frequency of the G allele=0.22), in high linkage disequilibrium (R2=0.83) with the insertion-deletion polymorphism identified in suicide attempt in bipolar disorder (Table 2). Weak evidence for an association was found in major depressive disorder (rs28591567, p=0.03, odds ratio for the G allele=1.11); the locus was not associated with suicide attempt in schizophrenia (p=0.67). No significant associations were identified in the meta-analysis between suicide attempt in bipolar disorder and schizophrenia (see Figure S5 in the online supplement). Manhattan plots and tables of the top results from all GWASs are provided in Figures S1–S5 and Tables S9–S14 of the online supplement).
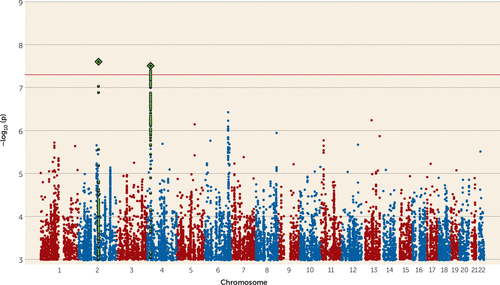
FIGURE 1. Manhattan plot from meta-analysis of genome-wide association studies on suicide attempt in major depressive disorder and bipolar disordera
a The red line illustrates the genome-wide significance threshold (p<5×10−8). SNPs in green are in linkage disequilibrium with the index SNPs (diamonds).
Replication Analysis
We tested SNPs from the three genome-wide significant loci for suicide attempt in the discovery phase for replication in independent mood disorder cohorts from the UK Biobank and iPSYCH. None of these loci showed association with suicide attempt in mood disorders in either study (minimum p value, 0.22) (Table 2; full results are shown in Table S15 in the online supplement).
Polygenic Risk Scoring and SNP Heritability
We performed polygenic risk scoring to investigate the genetic etiology of suicide attempt in major depressive disorder, bipolar disorder, and schizophrenia using scores from GWASs on psychiatric disorders and the suicide attempt GWASs conducted here. PRSs for major depression were significantly associated with suicide attempt in all three disorders (major depressive disorder: maximum variance explained on the liability scale, R2=0.25%, p=0.0006; bipolar disorder: R2=0.24%, p=0.0002; schizophrenia: R2=0.40%, p=0.0006) (Figure 2). The PRSs for bipolar disorder did not differ between suicide attempters and nonattempters, while PRSs for schizophrenia were significantly lower in suicide attempters compared with nonattempters (R2=0.42%, p=0.0005) (see Figure S6 in the online supplement).
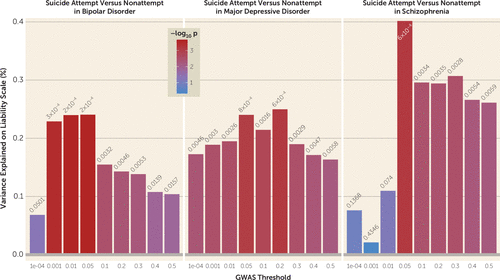
FIGURE 2. Higher polygenic risk scores for major depression in suicide attempters with bipolar disorder, major depressive disorder, or schizophrenia compared to nonattempters with the same disordersa
a The x-axis illustrates the p value threshold used to select SNPs from the discovery genome-wide association study (GWAS). The y-axis illustrates the percentage variance explained on the liability scale. The p values for the association between polygenic scores and suicide attempt are shown above each bar.
We used GREML to estimate the SNP heritability of suicide attempt in each psychiatric disorder. The h2SNP estimate of suicide attempt was 0.03 (SE=0.03, p=0.19) in major depressive disorder, 0.02 (SE=0.03, p=0.25) in bipolar disorder, and 0.10 (SE=0.07, p=0.06) in schizophrenia. None of these h2SNP estimates differed significantly from zero. Using these suicide attempt GWASs as discovery studies, we found that the PRSs for suicide attempt in a single psychiatric disorder were not associated with suicide attempt in another disorder (see Figure S7 in the online supplement).
Discussion
To our knowledge, this GWAS on suicide attempt is the largest performed to date, combining samples of suicide attempters and nonattempters across three major psychiatric disorders and 46 individual cohorts from the PGC. Three independent loci were associated with suicide attempt in the discovery phase. The strongest support was for the chromosome 4 locus in LOC105374524, a noncoding RNA, located approximately 500 kb downstream of the GBA3 (glucosylceramidase beta 3) gene and upstream of PPARGC1A, which is a transcriptional coactivator that regulates the genes involved in energy metabolism. This region reached genome-wide significance in the GWAS of suicide attempt in bipolar disorder, and the association strengthened in the meta-analysis of suicide attempt in mood disorders. Despite the support for this locus in the discovery phase, the association was not replicated in large independent mood disorder cohorts from the UK Biobank and iPSYCH. One possible explanation for this lack of replication is that more than 90% of suicide attempters in the replication cohorts had a depression diagnosis, while in the discovery PGC studies, this locus had the strongest effect in bipolar disorder. In addition, there may have been heterogeneity in the definition of suicide attempt, which in the discovery samples was based on psychiatric interviews and in the replication samples on self-report questionnaires and hospital records. For the other two loci that reached genome-wide significance for suicide attempt in the discovery phase, statistical power for replication was low, given the effect allele frequency of 2% at each locus.
The within-case analysis strategy used in this GWAS was designed to detect associations specific for suicide attempt and was informed by twin and family studies, which consistently indicate a genetic component of suicide attempt that is partially distinct from that of the psychiatric disorders themselves (5, 6). The LOC105374524 association with suicide attempt in mood disorders reported here has not been implicated in GWASs of major depression, bipolar disorder, or schizophrenia, indicating its specificity for suicide attempt and providing support for a partially distinct genetic etiology (18–20, 37, 38).
Through polygenic scoring analyses, we showed that genetic liability for major depression increases risk for suicide attempt; suicide attempters with major depressive disorder, bipolar disorder, or schizophrenia all had higher PRSs for major depression than nonattempters. Clinical studies are consistent with these findings, with increased depressive symptoms in bipolar disorder and the presence of depressive symptoms in schizophrenia being risk factors for suicide attempt (39, 40). Indeed, depressive symptom-based factor scores generated from clinical symptom data available for PGC schizophrenia case subjects were significantly higher in suicide attempters than in nonattempters (see the online supplement) (41). Our results suggest that the genetic etiology of suicide attempt may be partially unique and partially shared with major depression, and this finding is consistent with recent results showing high genetic correlation between these two phenotypes (13). Given the widespread pleiotropy between psychiatric disorders, some genetic overlap with other disorders is expected (42). To our knowledge, this study is the first to show that across disorders, suicide attempters carry a greater burden of depression risk alleles rather than simply a higher genetic liability for the psychiatric disorder they are affected by. The finding that nonattempters with schizophrenia have higher PRSs for schizophrenia, while suicide attempters with schizophrenia have higher PRSs for depression, demonstrates the genetic heterogeneity that exists within psychiatric cases. In previous studies, major depression PRSs have been shown to explain approximately 2% of variance in depression case-control status, compared with up to 0.4% of variance in suicide attempter status reported here (18). Despite the smaller variance explained, it is promising that PRSs can now be used in psychiatric cases to distinguish between individuals with different symptom profiles or comorbidities.
The SNP heritability estimates for suicide attempt in each disorder did not significantly differ from zero, and while several twin studies have reported that suicide attempt is moderately heritable, one study found that after adjusting for psychiatric disorders, the heritability decreased from 30% to 17% (5, 43). If this is a more accurate estimate of the independent genetic contribution to suicide attempt, then substantial increases in sample size will be required to fully interrogate its genetic etiology. Because estimated SNP heritabilities are generally much lower than twin heritabilities, our samples were insufficiently powered to identify this level of genetic contribution. Still, to our knowledge, the present study is the first consortium-based GWAS on suicide attempt and has made significant progress in increasing numbers by combining samples across clinical cohorts. Further collaborative efforts to amass samples on an even larger scale will be essential to achieve the success seen in GWASs of other psychiatric disorders. Data from population biobanks are now widely available, and leveraging these to conduct meta-analyses could rapidly increase statistical power. GWASs comparing suicide attempters with healthy control subjects may provide a complementary approach by prioritizing loci for follow-up in case-only studies, which should be expanded beyond studies of mood disorders and schizophrenia to include existing cohorts for other disorders in which suicide attempt is prevalent. The results of GWASs now robustly link hundreds of genetic loci to psychiatric disorders and provide additional opportunities to disentangle genetic effects on suicide attempt from the disorders themselves.
A strength of this study is that samples of suicide attempters have been successfully combined across many individual clinical cohorts. All 46 data sets were processed centrally using the same quality control, imputation, and analysis procedures. Suicide attempt and nonattempt were defined using items from structured psychiatric interviews. These items vary by interview, which may be considered a limitation, although the psychiatric interview or type of item used did not result in heterogeneity in the prevalence of suicide attempt across these cohorts (see the online supplement). Because study subjects were not ascertained primarily for suicide attempt, detailed information, such as the number of suicide attempts, medical consequences, or medication history, is not available for all participants. This study focused on lifetime suicide attempt to maximize the sample size, but some individuals who were nonattempters at recruitment may have attempted suicide later.
In conclusion, of all the psychiatric phenotypes, suicidality remains especially challenging to predict and assess, and there is an urgent need to better understand its etiology. As seen in GWASs of other psychiatric disorders, the number of genetic associations is expected to accumulate with increased sample size and can provide invaluable biological insights. Our novel finding that genetic liability for major depression increases risk of suicide attempt in major depressive disorder, bipolar disorder, and schizophrenia provides a possible starting point for predictive modeling and preventive strategies. The ultimate goal of genetic studies on suicide attempt is to translate these statistical associations into biological mechanisms and much-needed treatments and preventions for suicidality in order to reduce its burden on patients, families, and health care systems.
1 Preventing Suicide: A Global Imperative. Geneva, , 2014Google Scholar
2 Suicide: Facts at a Glance. Centers for Disease Control and Prevention, 2015. https://www.cdc.gov/violenceprevention/pdf/suicide-datasheet-a.pdfGoogle Scholar
3 : The impact of psychiatric illness on suicide: differences by diagnosis of disorders and by sex and age of subjects. J Psychiatr Res 2011; 45:1445–1452Crossref, Medline, Google Scholar
4 : Prevalence and comorbidity of mental disorders in persons making serious suicide attempts: a case-control study. Am J Psychiatry 1996; 153:1009–1014Link, Google Scholar
5 : Genetics of suicide: a systematic review of twin studies. Wien Klin Wochenschr 2007; 119:463–475Crossref, Medline, Google Scholar
6 : Family genetic studies, suicide, and suicidal behavior. Am J Med Genet C Semin Med Genet 2005; 133C:13–24Crossref, Medline, Google Scholar
7 : A genome-wide association study of attempted suicide. Mol Psychiatry 2012; 17:433–444Crossref, Medline, Google Scholar
8 : Genomewide association scan of suicidal thoughts and behaviour in major depression. PLoS One 2011; 6:
9 : Genome-wide association study of suicide attempts in mood disorder patients. Am J Psychiatry 2010; 167:1499–1507Link, Google Scholar
10 : Genetic relationships between suicide attempts, suicidal ideation and major psychiatric disorders: a genome-wide association and polygenic scoring study. Am J Med Genet B Neuropsychiatr Genet 2014; 165B:428–437Crossref, Medline, Google Scholar
11 : A genome-wide association study of suicidal behavior. Am J Med Genet B Neuropsychiatr Genet 2015; 168:557–563Crossref, Medline, Google Scholar
12 : Genomewide association studies of suicide attempts in US soldiers. Am J Med Genet B Neuropsychiatr Genet 2017; 174:786–797Crossref, Medline, Google Scholar
13 : Significant shared heritability underlies suicide attempt and clinically predicted probability of attempting suicide. Mol Psychiatry 2019Crossref, Medline, Google Scholar
14 : Genetics of suicide attempts in individuals with and without mental disorders: a population-based genome-wide association study. Mol Psychiatry 2018Crossref, Medline, Google Scholar
15 Diagnostic and Statistical Manual of Mental Disorders, 4th ed. Washington, DC, , 1994Google Scholar
16 International Classification of Diseases, 9th rev ed. World Health Organization, 1978Google Scholar
17 International Classification of Diseases, 10th rev ed. World Health Organization, 1992Google Scholar
18 : Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet 2018; 50:668–681Crossref, Medline, Google Scholar
19 : Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet 2019; 51:793–803Google Scholar
20 : Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511:421–427Crossref, Medline, Google Scholar
21 : Genotype imputation with thousands of genomes. G3 (Bethesda) 2011; 1:457–470Crossref, Medline, Google Scholar
22 : A linear complexity phasing method for thousands of genomes. Nat Methods 2011; 9:179–181Crossref, Medline, Google Scholar
23 : A map of human genome variation from population-scale sequencing. Nature 2010; 467:1061–1073Crossref, Medline, Google Scholar
24 : Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015; 4:7Crossref, Medline, Google Scholar
25 : Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38:904–909Crossref, Medline, Google Scholar
26 : METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26:2190–2191Crossref, Medline, Google Scholar
27 : PRSice: polygenic risk score software. Bioinformatics 2015; 31:1466–1468Crossref, Medline, Google Scholar
28 : A better coefficient of determination for genetic profile analysis. Genet Epidemiol 2012; 36:214–224Crossref, Medline, Google Scholar
29 : GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011; 88:76–82Crossref, Medline, Google Scholar
30 : Genetic power calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 2003; 19:149–150Crossref, Medline, Google Scholar
31 : Statistical power to detect genetic (co)variance of complex traits using SNP data in unrelated samples. PLoS Genet 2014; 10:
32 : Power and predictive accuracy of polygenic risk scores. PLoS Genet 2013; 9:
33 : A fast method that uses polygenic scores to estimate the variance explained by genome-wide marker panels and the proportion of variants affecting a trait. Am J Hum Genet 2015; 97:250–259Crossref, Medline, Google Scholar
34 : UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015; 12:
35 : Genome-wide genetic data on ∼500,000 UK Biobank participants. bioRxiv 2017.
36 : The iPSYCH2012 case-cohort sample: new directions for unravelling genetic and environmental architectures of severe mental disorders. Mol Psychiatry 2018; 23:6–14Crossref, Medline, Google Scholar
37 : Genome-wide meta-analysis of depression in 807,553 individuals identifies 102 independent variants with replication in a further 1,507,153 individuals. bioRxiv 2018.
38 : Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet 2018; 50:381–389Crossref, Medline, Google Scholar
39 : Prospective predictors of suicide and suicide attempts in 1,556 patients with bipolar disorders followed for up to 2 years. Bipolar Disord 2006; 8:566–575Crossref, Medline, Google Scholar
40 : Schizophrenia and suicide: systematic review of risk factors. Br J Psychiatry 2005; 187:9–20Crossref, Medline, Google Scholar
41 : Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol Psychiatry 2014; 19:1017–1024Crossref, Medline, Google Scholar
42 : Analysis of shared heritability in common disorders of the brain. Science 2018; 360:
43 : A twin study of genetic and environmental influences on suicidality in men. Psychol Med 2002; 32:11–24Crossref, Medline, Google Scholar



