Clinically Useful Genetic Markers of Antidepressant Response: How Do We Get There From Here?
Predicting who will respond to an antidepressant—and who will not—remains an urgent but unfulfilled need in psychiatry (1). Clinical indicators of response have a long history, but probably have low predictive value. This state of affairs has fueled an energetic search for biomarkers.
Genetic markers are ideal biomarkers, since they do not suffer from reverse causation. Reverse causation arises when a biomarker value is actually the consequence of the outcome, rather than its cause. Genetic markers that represent the inherited sequence of DNA cannot reflect the result of a health outcome. The best genetic markers would identify individuals most likely to benefit from a particular treatment and least likely to suffer adverse events. Years of work with candidate genes have yielded some reproducible findings, but their individual effects are small, with little impact on clinical decision making (2). Studies that use genome-wide genotyping technology have implicated additional genes (3), but well-replicated findings that can help guide clinical decision making remain elusive.
In this issue, Schatzberg et al. (4) report that common genetic markers in ABCB1 provide some information about which patients are most likely to respond to certain selective serotonin reuptake inhibitors. ABCB1 encodes P-glycoprotein, which influences the efficiency with which certain antidepressants (and other drugs) cross the blood-brain barrier. Common variants in ABCB1 have been associated with antidepressant outcome in some previous studies (5), so replication in an independent sample was the next logical step. Ten common markers in and around ABCB1 were tested in 683 patients with major depressive disorder who were treated for at least 2 weeks and an additional 576 patients who were treated for 8 weeks as part of the International Study to Predict Optimized Treatment in Depression (iSPOT-D) trial. The samples were carefully ascertained and evaluated, with prospective measures of efficacy and side effect burden, using standard instruments. General and emotional cognition were further assessed with a battery of 13 tests. Antidepressants included escitalopram, sertraline, and extended-release venlafaxine, all of which are known to bind P-glycoprotein. One single-nucleotide polymorphism (SNP) upstream of ABCB1 was associated with remission and subjective side effects, and the direction of the effect differed among the medications tested.
How far does this take us, and what’s next? Treatment assignment was randomized, but not blinded, and there were no placebo controls and no direct assessments of adherence. Despite these limitations, this study represents one of the few replications in the field of antidepressant pharmacogenomics, and is thus a real step forward. Still, the effect sizes are too small to be useful in clinical decision making.
It seems likely that antidepressant response, like depression itself, is a polygenic trait, influenced by numerous genes, each of small effect. Therefore, many more genetic markers will be needed before we can achieve clinically significant prediction. This will require much larger samples than have been studied so far. We now know that genome-wide association studies (GWAS) require very large sample sizes to identify genetic markers of polygenic traits. Samples in the tens of thousands are now routine, yet no GWAS of antidepressant outcome to date has exceeded 2,000. As pharmacogenomics begins to bear fruit in psychiatry, we can hope for a renewed focus on the large sample sizes necessary for robust findings.
The heritability of antidepressant response puts an upper limit on what can be predicted by genetic markers alone. Heritability measures how much of the individual differences in a trait can be explained by inherited genetic differences. This is typically estimated from twin studies, but these are generally impractical for treatment response studies, since twins would only rarely be treated for the same illness in the same way during the time frame of a study. An alternative approach directly compares genetic similarity (estimated from a genome-wide set of SNPs) with phenotypic similarity using sophisticated regression methods (6). One study used this approach to estimate that antidepressant response was about 40% heritable, but with a large confidence interval (7). Other statistical models show that traits with this degree of heritability cannot be predicted very well from genetic markers alone (8). Even a panel of genetic markers that explains a quarter of the genetic variance—more than any GWAS to date—gives an area under the curve of only about 64%. Thus, a combination of clinical data and genetic and other biomarkers will be needed if we are to push predictability into the clinically useful range.
Advances in cancer chemotherapy may light the way. We can already tailor treatment for breast cancer based on a combination of family history, clinical staging, and expression of estrogen receptors by tumor cells. Can we not envision an analogous combination of variables that would identify patients most likely to respond to antidepressants (Figure 1)? Studies like that of Schatzberg et al. are encouraging because they suggest that the ultimate goal of clinically useful outcome prediction can be achieved.
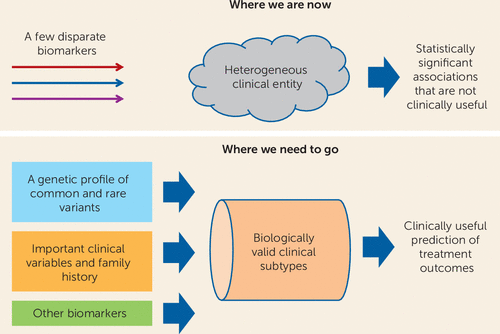
FIGURE 1. Where We Are Now and Where We Need to Goa
a Current data support statistically significant associations with treatment outcomes, but these are not clinically useful. These first steps suggest that more progress can be made if we can develop models that incorporate clinical, genetic, and other biomarker data that can be applied to more biologically valid clinical subtypes of depression.
Further progress may depend, in part, on the development of better ways to break down the highly heterogeneous clinical entity of, say, “major depression” into more homogeneous subgroups with more shared underlying biology. Clinical variables may play a role, but it is limited. Also in this issue of the Journal, Arnow et al. (9), in another iSPOT-D report, show that the traditional clinical subtypes of major depression seem to play little or no role in predicting response to antidepressant treatment. Other common-sense clinical variables, such as severity, recurrence, and early onset may have some value but so far have not been tested in large samples. Genetic and other biomarkers will clearly be needed, but so far GWAS have not yielded robust, genome-wide significant findings in major depression. Rarer genetic variation, which is not well tested by GWAS, could play a role in a minority of individuals, as has been seen in studies of rare copy number variants in schizophrenia, autism, and bipolar disorder (10). Patients with treatment-resistant illness may harbor rare variants that block response to standard treatments. Studies of other biomarkers have provided inconsistent results, but some neuroimaging measures (11) show promise. Markers of epigenetic changes are also likely to play an important role (12).
We also need studies that look beyond the still quite narrow pharmacology of currently labeled antidepressants to other agents and treatment modalities. Could genetic markers help identify patients who are more likely to respond to unconventional antidepressants, such as ketamine, or to neuromodulatory treatments, such as ECT? Early identification of patients who are unlikely to respond to antidepressant medications alone could greatly reduce the burdens of chronicity and may reduce the proportion of patients whose illness develops into treatment-resistant depression or suicide.
Further progress will require continued investments in public research funds and within industry. But we should also take a renewed look at the potential presented by the many thousands of patients who undergo routine antidepressant therapy every year. We need better ways to track outcomes and integrate this information with genetic and other data. New models of research consent will clearly be needed, so that more patients have the opportunity to participate in research while undergoing routine care in hospitals and outpatient settings. Here again, our colleagues in the cancer field may offer some useful examples. Just as most chemotherapy patients are enrolled in various kinds of outcome studies, so that something is learned from every patient, regardless of outcome, psychiatric patients could also be offered the opportunity to advance the field while receiving the best treatment we have to offer. We’ve made a start, but we can go much further. Anything less is a lost opportunity both for psychiatry and for the patients we serve.
1 : Pharmacogenomics and personalized medicine in neuropsychiatry. Neuron 2012; 74:773–776Crossref, Medline, Google Scholar
2 : Review and meta-analysis of antidepressant pharmacogenetic findings in major depressive disorder. Mol Psychiatry 2010; 15:473–500Crossref, Medline, Google Scholar
3 : Genome-wide association studies in pharmacogenomics of antidepressants. Pharmacogenomics 2015; 16:555–566Crossref, Medline, Google Scholar
4 : ABCB1 genetic effects on antidepressant outcomes: a report from the iSPOT-D trial. Am J Psychiatry 2015; 172:751–759Link, Google Scholar
5 : Polymorphisms in the drug transporter gene ABCB1 predict antidepressant treatment response in depression. Neuron 2008; 57:203–209Crossref, Medline, Google Scholar
6 : Estimation of SNP heritability from dense genotype data. Am J Hum Genet 2013; 93:1151–1155Crossref, Medline, Google Scholar
7 : Contribution of common genetic variants to antidepressant response. Biol Psychiatry 2013; 73:679–682Crossref, Medline, Google Scholar
8 : Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res 2007; 17:1520–1528Crossref, Medline, Google Scholar
9 : Depression subtypes in predicting antidepressant response: a report from the iSPOT-D trial. Am J Psychiatry 2015; 172:743–750Link, Google Scholar
10 : Rare copy number variation in treatment-resistant major depressive disorder. Biol Psychiatry 2014; 76:536–541Crossref, Medline, Google Scholar
11 : Toward a neuroimaging treatment selection biomarker for major depressive disorder. JAMA Psychiatry 2013; 70:821–829Crossref, Medline, Google Scholar
12 : Epigenetic mechanisms of depression and antidepressant action. Annu Rev Pharmacol Toxicol 2013; 53:59–87Crossref, Medline, Google Scholar



