G72 and Its Association With Major Depression and Neuroticism in Large Population-Based Groups From Germany
Abstract
Objective:G72 is among the most frequently replicated vulnerability genes for schizophrenia and bipolar disorder. The authors previously found identical haplotypes of markers M23 and M24 to be associated with schizophrenia, bipolar disorder, and panic disorder. Given both the well-recognized familial clustering across these disorders and recent linkage findings implicating the region harboring G72 in the etiology of major depression and panic disorder, we can hypothesize that G72 should also be involved in the etiology of major depression. Neuroticism, measuring trait anxiety, may be the endophenotypic link underlying genetic associations with G72 across diagnostic boundaries. The authors tested whether the previously observed risk haplotypes are also associated with major depression and neuroticism. Method: The authors performed a standard haplotype analysis in a group of 500 major depression patients and 1,030 population-based comparison subjects. The authors also performed an exploratory analysis on 10 additional G72 markers using a novel haplotype-sharing approach. They performed a quantitative trait haplotype analysis in an independent group of 907 individuals phenotyped for neuroticism. Results: The previously identified M23-M24 risk haplotype was significantly associated with major depression and high levels of neuroticism. The haplotype-sharing analysis also implicated the same region, whereas more proximal markers showed no association with major depression. Conclusions: This is the first study to the authors’ knowledge to implicate the G72 locus in the etiology of major depression and neuroticism. The results strengthen the notion of a genetic overlap between diagnoses, commonly conceptualized as distinct entities. Neuroticism may constitute the common underlying endophenotypic link.
Since the initial report by Chumakov et al. (1) of an association between markers at the G72 locus and schizophrenia in samples from Russia and Canada and subsequent reports by Hattori et al. (2) , Chen et al. (3) , and Schumacher et al. (4) showing association between this locus and bipolar disorder in samples from the United States and Germany, this locus has become the focus of increased attention in psychiatric genetic research. A recent meta-analysis by Detera-Wadleigh and McMahon (5) concluded that the association findings between G72 and both schizophrenia and bipolar disorder are “among the most compelling in psychiatry” despite the fact that associated alleles and haplotypes are not identical across studies and functional variants remain to be demonstrated. Findings are not limited to a specific population, being observed in European, North American, Han Chinese, and Ashkenazi samples, with few studies reporting nonassociations (5 – 9) . Our group reported association of identical G72 haplotypes with both schizophrenia and bipolar disorder in the German population (4) . In line with findings from other groups on G72 , our findings support the notion of a genetic overlap across major psychiatric diagnoses. Such a genetic overlap has also been suggested by studies of DTNBP1 (dysbindin), COMT, BDNF, DISC1 , and NRG1(10 – 17) .
Although there is now increasing support for the G72 locus as a susceptibility gene for schizophrenia and bipolar disorder (15 – 18) as well as evidence for an etiological role in panic disorder (19) , a potential association with major depression still needs to be assessed, in particular given the well-established familial clustering of major depression, schizophrenia, bipolar disorder, and anxiety disorders (20 – 22) . Moreover, a large multicenter study provided evidence of linkage between major depression and a locus on 13q31.1–q31.3 (76.1–92.6 megabases on National Center for Biotechnology Information build 35), thus lying in the vicinity of G72 (104.9 megabases on National Center for Biotechnology Information build 35; reference 23 ).
To pursue this issue, we undertook a two-tiered design. First, we tested whether the previously identified susceptibility haplotypes for schizophrenia, bipolar disorder, and panic disorder, i.e., the M23-M24 haplotypes C-T and T-A, were also associated with major depression with a group of 500 major depression patients and 1,030 population-based comparison subjects from Germany, all of German descent. Second, in addition to standard haplotype analyses focusing on the previously reported haplotypes, we also conducted an exploratory analysis with an additional 10 single nucleotide polymorphisms (SNPs) and a novel haplotype-sharing approach. This step was performed in order to obtain a complete overview of the contribution of genetic variability at the G72 locus to major depression.
Finally, in order to facilitate a better understanding of why susceptibility genes such as G72 are consistently found associated across diagnostic boundaries, we took our analysis a step further by studying a potential association between the same susceptibility haplotypes and neuroticism in 907 individuals from the general population. The rationale for this approach is based on the large body of evidence that neuroticism can be considered a predictor and potential endophenotype for several psychiatric disorders, including major depression and schizophrenia (24 – 28) and, on the other hand, follows from our previously formulated hypothesis that the association between G72 and major psychiatric phenotypes may be due to an association with an underlying trait of anxiety (29) .
Method
Groups, Recruitment, and Phenotype Characterization Procedures
Cases were recruited from consecutive admissions to the inpatient units of the Department of Psychiatry and Psychotherapy of the University of Bonn, Germany. DSM-IV lifetime diagnoses of major depression were made by a consensus best-estimate procedure (30) , based on all available information, including a structured interview (the Structured Clinical Interview for DSM-IV [31] ), medical records, and the family history method. We also used the OPCRIT (32) system to obtain detailed polydiagnostic documentation of symptoms. The details of our recruitment and phenotype characterization procedures are outlined elsewhere (33 , 34) .
For the analyses, we included 500 major depression patients, 178 men (36%) and 322 women (64%). As regards the clinical manifestation, 124 were diagnosed with single episode of major depression and 376 with a recurrent major depression. Four hundred patients (80.0%) had a melancholic subtype. As regards a positive family history in first- or second-degree relatives, 69 (13.8%) patients had a family history of major depression, five (1.0%) had a family history of bipolar disorder, and 16 (3.2%) had a family history of schizophrenia. The mean (SD) age at assessment was 47.9 (SD=13.8) years; the mean (SD) age at onset was 36.9 (SD=13.3) years, age at onset being defined as the time at which criteria for a DSM-IV major depression were met for the first time.
The population-based comparison group was established with the support of the local census bureau of the city of Bonn, Germany (North Rhine-Westphalia). For the analyses, we included 1,030 (499 men, 531 women) individuals; the mean (SD) age at assessment was 47.9 (SD=15.5) years. This group was established within the framework of the German National Genome Research Network I (Nationales Genomforschungsnetz I, www.ngfn.de) of the Federal Ministry of Education and Research (www.bmbf.de) between 2000 and 2003 to serve as an epidemiological comparison group for complex genetic studies within the NGFN.
All comparison subjects were seen by a trained psychiatrist and screened for psychiatric disorders, as described previously (34) . From the 1,030 individuals, 133 (13%) had a history of major depression, one (0.09%) of schizophrenia, four (0.4%) of bipolar disorder, and 14 (1.3%) of anxiety disorder. To maintain the character of an epidemiological community group and minimize type II errors, we opted to keep all 1,030 individuals as comparison individuals in our primary analysis. All cases and the comparison individuals were of German descent, which, in our study, is fulfilled when an individual’s parents originate from Germany and when there is no indication that one of the four grandparents may originate from outside of Germany.
General Population-Based Group for the Study of Neuroticism
We recruited 907 people (430 men, 477 women; mean age=28.8 years, SD=11.3) from the general population of the Rhineland region at three research centers (Aachen, Trier, and Mannheim). Neuroticism scores were obtained with the established and widely used NEO-FFI questionnaire (35) . Neuroticism scores were transformed to follow a normal distribution with a mean value of 100 and an SD of 10.
Ethical Considerations
Protocols and procedures were approved by the institutional review boards ( Ethikkommission ) of the respective academic institutions. After complete description of the study to the subjects, written informed consent was obtained.
Genotyping
Major depression cases and comparison subjects were genotyped for 12 G72 SNPs ( Table 1 ) with the MassARRAY system (Sequenom Inc., San Diego). The two markers M23 and M24 were the subject of our primary analysis testing the specific hypothesis that their two-marker haplotypes, C-T and T-A, were associated with major depression. The remaining 10 SNPs were analyzed within our exploratory analyses as described below. Of these, SNPs M12 through rs1935062, M21, and M22 were chosen on the basis of previously reported association findings (5) . DAO_3UTR_SNP12 was only very recently found to be associated with bipolar disorder and major mood episodes in schizophrenia by Williams et al. (14) . Marker G72_z6:1117 was identified by resequencing in our laboratory. Overall, the 12 markers cover a ∼95 kilobase region, including the 5′ and 3′ flanking regions of the G72 locus, constituting the region of interest for linkage disequilibrium mapping (5) . Genotyping completeness ranged from 96.3% (marker rs1935062) to 99.1% (marker rs1421292/M24). The group studied for neuroticism was only typed for markers M23 and M24.
For quality comparison purposes, we genotyped a subset of the group in duplicate in order to estimate the replicate error rate. Two of 96 DNA samples were randomly chosen for this purpose. For the SNPs genotyped in the course of this study, all genotypes between duplicates were consistent (0% replicate error rate). Apart from that, positive and negative controls are always included routinely in our genotyping experiments.
By a standard 1 df chi-square test, there were no significant deviations from Hardy-Weinberg equilibrium for the genotype distributions of the studied groups.
Calculation of Linkage Disequilibrium
Intermarker linkage disequilibrium, as expressed by D′, was calculated and visualized using the HAPLOVIEW (http://www.broad.mit.edu/mpg/haploview/contact.php; reference 36 ) software. Linkage disequilibrium was calculated in the comparison group (N=1,030).
Primary Statistical Analysis on Major Depression
Case-control haplotype analyses for the two-marker haplotype, consisting of markers M23 and M24, were performed with version 3.0 of the widely used program UNPHASED (http://www.mrc-bsu.cam.ac.uk/personal/frank/software/#software; reference 37 ). Using a standard unconditional logistic regression, UNPHASED performs likelihood ratio tests under a log-linear model of the probability that a haplotype belongs to a case rather than a comparison subject; the expectation-maximization algorithm is used to resolve uncertain haplotypes and provide maximum-likelihood estimates of frequencies. Based on our previous findings in schizophrenia, bipolar disorder, and panic disorder (4 , 19) , we specifically tested the M23-M24 haplotypes C-T and T-A. Empirical p values were established by performing 10,000 permutations.
Secondary Statistical Analysis on Major Depression
Three exploratory analyses were carried out: first, we tested whether we would obtain similar results for the M23-M24 haplotypes when the comparison group was restricted to the 871 individuals from the community group without a lifetime history of a psychotic, affective, or anxiety disorder. Second, we tested whether confining case definition to recurrent major depression (N=376) would yield a similar effect. Third, we wanted to obtain a more comprehensive overview of the contribution of genetic variability in the G72 region to major depression, besides the specific analysis of the region captured by markers M23 and M24. To achieve this, we opted for a novel haplotype-sharing method (38) in order to exhaust the joint information conferred by all the markers in the critical region.
Haplotype-sharing analysis is based on the assumption that, in the neighborhood of a genetic susceptibility variant, haplotypes carrying this variant (“case” haplotypes) are more related than haplotypes not carrying the mutation (“random” or “comparison” haplotypes) and that the time to the most recent common ancestor is shorter among case haplotypes than among random haplotypes. It is thus expected that case haplotypes share significantly longer stretches of DNA “identical by descent” around the variant because of fewer recombinational and mutational events (39 – 42) . Haplotype-sharing analysis was first successfully applied in the study of monogenic disease (41 , 42) . In the last decade, it has furthermore become an established method in the analysis of complex disorders (39 , 43 – 49) . Here, we have applied the approach of Mantel’s statistics for space-time clustering (50) to correlate genetic and phenotypic similarity, as developed by Beckmann et al. (38) : where x denotes a putative disease locus and i and j are haplotypes. The sum is over all pairwise comparisons of haplotypes. The genetic similarity, Lij(x) , is measured as the number of intervals surrounding x flanked by markers identical by state (haplotype sharing). Ysisj denotes the phenotypic similarity of the individuals si and sj and is defined as the mean-corrected product Ysisj =(y si– μ y )(y sj– μ y ), where μ y denotes the sample mean and ysi and ysj the disease status of si and sj . In the case-control scenario, ysi is 1 if si is affected and 0 if si is a comparison individual. Thus, the Mantel statistics contrast between the pairwise comparisons of cases versus cases, cases versus comparison subjects, and comparison subjects versus comparison subjects. The statistical significance is evaluated by a Monte Carlo permutation in which the phenotype Y is permuted randomly over the individuals, while keeping together the two haplotypes derived from the same individual. Although the Mantel statistics use the information of multilocus haplotypes, they are pointwise statistics. The Mantel statistics have been applied in the software TOMCAT (www.dkfz.de/en/klepidemiologie/software/software.html). For this analysis, individual haplotype pairs were estimated with the software FASTPHASE (51) .
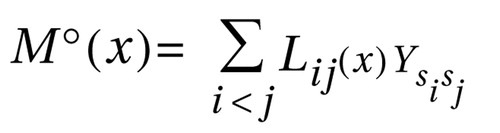
Correction for Multiple Testing in the Haplotype-Sharing Analysis
To correct for multiple testing in the exploratory haplotype-sharing analysis, a step-down min-p algorithm was used to adjust the p values according to the number of tests, i.e., the number of SNPs. This algorithm has been described in detail by Obreiter et al. (52) and has been implemented in the software program SDMinP (52) . SDMinP is suited for the fast calculation of empirical and adjusted p values for correlated and uncorrelated hypotheses in multiple testing experiments. It is based on the free step-down resampling method for controlling the familywise error rate.
Quantitative Trait Association Study of Neuroticism
In analogy to the case-control study on major depression, association between neuroticism and the M23-M24 haplotypes C-T and T-A was tested using UNPHASED version 3.0 (37) .
Post Hoc Power Analyses
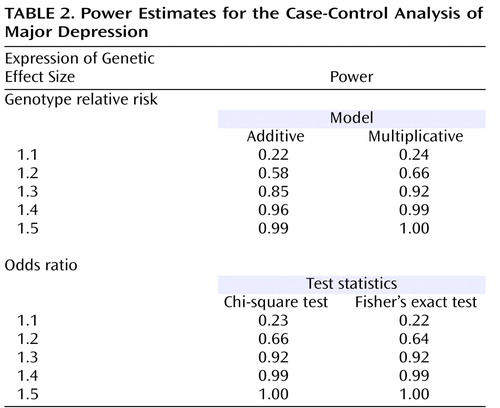
Power analyses for the case-control analysis on major depression were performed with the Genetic Power Calculator (http://pngu.mgh.harvard.edu/∼purcell/gpc/; reference 53 ). Power estimates are based on a marker allele frequency of 0.5 (e.g., reflecting the frequencies of the M23-M24 haplotypes) and an alpha level of 5%. Power was calculated both under an additive and a multiplicative model, assuming varying degrees of genotype relative risk. We furthermore assumed a risk allele frequency of 0.5 and complete linkage disequilibrium (D′=1) between marker and risk allele. Power was also estimated expressing the genetic effect size as an odds ratio with the PS Power and Sample Size Calculations suite of programs (http://www.mc.vanderbilt.edu/prevmed/ps/index.htm; reference 54 ). The genetic power calculator (53) was also used to determine the power for the quantitative trait analysis on neuroticism. We estimated power for total quantitative trait locus variances of 1%, 2%, and 3% and no dominance variance. We furthermore assumed a quantitative trait locus increaser allele frequency of 0.5 and complete linkage disequilibrium (D′=1) between marker and quantitative trait locus increaser allele.
Results
Patterns of Linkage Disequilibrium
Altogether, 12 G72 markers were studied (rs numbers, trivial names, and physical positions are presented in Table 1 ). The patterns of intermarker linkage disequilibrium are as depicted in Figure 1 . Patterns of linkage disequilibrium (expressed in D′) are similar to the patterns in a trio sample of European origin from the HapMap data (5) and a sample from the United Kingdom (14) . The linkage disequilibrium structure is characterized by two blocks of high linkage disequilibrium between markers M12 and DAO_3UTR_SNP12 and between markers M22 and M24, respectively, with marker M21 mapping to a region of low linkage disequilibrium.
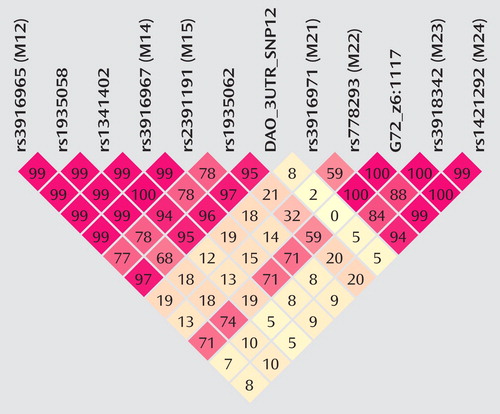
a The value of intermarker linkage disequilibrium for any given pair of markers (also see Table 1) is given in the respective square. A two-digit value, e.g., 97, stands for a D′ value of 0.97; a value of 2 for 0.02, and 100 stands for a value of 1.00. High levels of linkage disequilibrium are represented by red coloring, with decreasing color intensity representing lower levels of linkage disequilibrium.
Primary Statistical Analysis: Case-Control Study of Major Depression
Cases and comparison subjects were assessed for a potential differential distribution of the C-T and the T-A haplotypes. C-T was more frequent in cases than in comparison subjects (53.1% versus 49.4%), whereas the T-A haplotype was less frequent in cases than in comparison subjects (43.8% versus 48.1%), conferring an odds ratio of 1.18, (95% confidence interval [CI]=1.01–1.38, p=0.04, permuted p<0.05). Given that the gender ratios differed between major depression cases and comparison subjects, we further tested for a differential haplotype distribution between men and women in the combined group of cases and comparison subjects; this was not the case (p=0.64). Thus, our result is unlikely to be due to stratification by gender. Individual allele frequency differences between cases and comparison subjects are given in Table 1 .
Secondary Statistical Analysis
Community comparison group
When the aforementioned case-control analysis was performed with only the 871 community comparison individuals without a lifetime history for a psychotic, affective, or anxiety disorder, the results remained virtually unchanged: comparison of major depression cases with comparison subjects also showed a differential distribution of the M23-M24 haplotypes. The C-T haplotype was more frequent in cases than in comparison subjects (53.1% versus 49.5%), whereas the T-A haplotype was less frequent in cases than in comparison subjects (43.8% versus 48.2%). The associated odds ratio was 1.18 (95% CI=1.01–1.38). Owing to the 25% smaller size of the comparison group, the observed p value is slightly larger than for the primary analysis comprising the full set of comparison subjects (p<0.05, permuted p=0.053).
Case definition confined to recurrent major depression
Similar to the aforementioned secondary analysis, the decrease in group size, from 500 to 376 cases, also led to an increase in the observed significance level: the C-T haplotype was more frequent in cases than in comparison subjects (52% versus 49.4%), whereas the T-A haplotype was less frequent in cases than in comparison subjects (44.8% versus 48.1%). The associated odds ratio was 1.14 (95% CI=0.96–1.35, p=0.15, permuted p=0.16).
Haplotype sharing
Guided by the linkage disequilibrium structure of the locus and the physical intermarker distances, haplotype sharing was not evaluated for the complete 94.8 kilobase range defined by all 12 markers but separately for intervals described by markers M12 through DAO_3UTR_SNP12 and M21 through M24. Correction for multiple testing was based on these intervals.
Markers M21, M23, and M24 (p=0.06 for all) fell short of nominal levels of significance. The neighboring markers M22, p=0.02) and G72_z6:1117 (p=0.04) were significantly associated with major depression. The proximal markers (M12 through DAO_3UTR_SNP12) showed no association at all. All p values have been adjusted for multiple testing, as described ( Figure 2 ).

a The pointwise p values obtained from the haplotype-sharing analyses and corrected for multiple testing are plotted. Empirical p values were derived by 10,000 Monte Carlo permutations. From left to right, the diamonds represent the 12 markers as listed in Table 1. Corrected p values are plotted in logarithmic transformation and correct relative physical marker positions are given. The horizontal line indicates a nominal level of significance (p=0.05).
Quantitative Trait Association Study of Neuroticism
A significant association was observed between neuroticism and the tested M23-M24 haplotypes. C-T was significantly associated with higher neuroticism scores than T-A (p<0.02, permuted p<0.02). The additive value of C-T versus T-A, i.e., the change in expected mean trait value due to this haplotype, was 0.95.
Post Hoc Power Analyses
Detailed power estimates for the case-control analysis on major depression are presented in Table 2 . In summary, our group of 500 cases and 1,030 comparison subjects had sufficient power (>0.8) to detect an effect at a genotype relative risk or odds ratio of 1.3 or larger. For the quantitative trait analysis on neuroticism, power estimates were as follows: assuming total QTL variances of 1%, 2%, and 3%, power estimates were 0.67, 0.92, and 0.99, respectively.
Discussion
Since its initial description (1) , the G72 locus has become one of the most frequently replicated susceptibility genes for both schizophrenia and bipolar disorder (5 , 16 – 19) . In our previous work, we found identical G72 alleles and haplotypes to be associated with schizophrenia, bipolar disorder, and panic disorder in independent samples from Germany (4 , 19) . Thus, we demonstrated for the first time the involvement of G72 variants in three distinct clinical diagnoses.
Familial clustering across mood, anxiety, and psychotic disorders is a well-established finding in psychiatry (20 – 22) . Findings from a large multicenter study provided evidence of linkage between major depression and a locus on 13q in proximity to G72(23) . Furthermore, 13q has also been implicated in the etiology of panic disorder (55) . This prompted us to test whether G72 was also involved in the etiology of major depression with a two-tiered design. Our primary analysis tested the specific hypothesis that the M23-M24 haploytpes C-T and T-A that we had previously found associated with schizophrenia, bipolar disorder, and panic disorder were also associated with major depression. With a large group of patients with major depression, we were able to identify an association between major depression and the M23-M24 haplotypes C-T and T-A, with C-T being more frequent and T-A being less frequent in cases. This finding was furthermore corroborated by the haplotype-sharing analysis, implicating an involvement of the distal region of G72 in the etiology of major depression, with markers M22 and G72_z6:117 reaching significance after correction for multiple testing. Markers M23 and M24 did not reach significance (p=0.06) after correction.
The fact that the standard haplotype analysis with UNPHASED and the haplotype-sharing analysis do not completely overlap in their results illustrates that these two approaches are fairly independent from each other because haplotype information is considered in different ways. At every marker position, the Mantel statistic tests whether there is an excess of sharing between case and comparison haplotypes around this SNP. Thus, it is a pointwise test taking into account the information of haplotypes to calculate the shared length as a measure of similarity. In contrast, UNPHASED tests for differences in haplotype frequencies between cases and comparison subjects in a logistic regression framework. However, given that the two identified sets of SNPs (haplotype-sharing analysis: M22 and G72_z6:1117; standard haplotype analysis: M23 and M24) are very close to each other, i.e., less than 10 kilobases, one can assume that both methods do identify the same region. The difference between the two approaches also explains why the haplotype-sharing analysis may highlight an SNP showing similar allele frequencies for cases and comparison subjects. The selection of the 12 G72 markers was not based on a systematic HAPMAP-based coverage of the region, but rather, we took into account the comprehensive association findings at the G72 locus and investigated the potential effect of those markers that had shown the most promising results with psychiatric phenotypes in previous studies.
Taking the results of the present study and the findings from our previous studies, there is now evidence for an association of G72 with four major diagnostic entities: schizophrenia, bipolar disorder, panic disorder, and major depression. These association findings were obtained for identical haplotypes, i.e., the T-A and the C-T haplotypes of markers M23 and M24, in one of the largest sample collections worldwide, including more than 3,000 individuals from German and Polish populations (4 , 19 , 29) .
The association between this locus and major psychiatric disorder is still surrounded by some important caveats, however, given that the association findings with schizophrenia and bipolar disorder and with specific subtypes, e.g., schizophrenia with mood episodes or bipolar disorder with psychotic features (14 , 29) , have been obtained with different alleles, even in similarly sized and phenotyped samples of European origin, which may be due to a variety of reasons, ranging from so far undetected ethnic stratification to complex expression control (5 , 56 , 57) . We therefore acknowledge that evidence remains that other G72 variants play an etiological role. However, since this situation is not unique to G72 but also affects other vulnerability genes for psychiatric disorders such as dysbindin (58) , we and others (M. O’Donovan and N. Craddock, American College of Neuropsychopharmacology meeting, December 2006) believe that it should not prevent further research in these loci, in particular, as the situation of different alleles being associated across studies may be consistent with so far undetected locus-locus interactions and subtle differences in the linkage disequilibrium structure (59) . In contrast, we strongly advocate the continuation of this line of research. Identifying the functional variants of G72 and other vulnerability genes will necessitate the refinement of both psychiatric phenotypes and molecular genetic research techniques. This line of research should be accompanied by further studies on the biological relevance of G72(1 , 60) .
We further acknowledge that the genetic effect sizes we obtained for the individual M23-M24 susceptibility haplotypes are very modest. However, these odds ratios are within the range for reported associations with schizophrenia and bipolar disorder (5 , 56) and are typical for the situation in complex disorders (61) . Moreover, they are consistent with the notion of a polygenic etiology of complex disorders, such as psychiatric phenotypes, a concept that is gaining increased attention (62 – 65) . For diabetes (66 , 67) and, very recently, for bipolar disorder (68 , 69) , polygenic etiologies are supported by large-scale genome-wide association studies. In other words, the G72 risk haplotypes that we have found associated with major depression are not likely to play a major role in the pathophysiology of this disorder when considered alone. However, within the context of a polygenic model, each person’s disease risk is influenced by the total burden of risk alleles or haplotypes they carry; taken for themselves alone, these alleles or haplotypes only confer very modest effects. Disease occurs when the burden of alleles or haplotypes crosses some threshold.
The major strength of our study lies in its large group size and robust recruitment and phenotyping procedures. A study with a smaller group size and less stringent methods may have missed the modest effect we describe (34) , in particular for an etiologically heterogeneous disorder like major depression (70 , 71) .
The challenge lying ahead is to dissect this heterogeneity. This will facilitate a better understanding of why susceptibility genes such as G72 are consistently found associated across diagnostic boundaries. Ideally, one would want to pinpoint an (endo)phenotype linking different diagnostic entities together (72 , 73) . For the case of G72 , we could previously show that persecutory delusions constituted the common link between the association findings for schizophrenia and those for bipolar disorder in samples from Germany and Poland (29) . Given our G72 findings on panic disorder, the evidence for linkage between panic disorder and chromosomal region 13q (55) , where G72 resides, and the notion that trait anxiety or worry are important predictors for the development, intensity, and persistence of persecutory delusions (74 , 75) , we have furthermore advocated that not persecutory delusions per se but rather a trait of anxious affectivity might constitute this common link.
Therefore, in the present study, we set out to test this hypothesis by studying a potential association between G72 and trait anxiety, i.e., neuroticism. In a large group of 907 individuals from the general population of the Rhineland region, we detected association between the M23-M24 risk haplotypes and neuroticism. In concordance with the findings on schizophrenia, bipolar disorder, panic disorder, and now major depression, in which the C-T haplotype was associated with an increased disease risk, the very same haplotype was associated with higher levels of neuroticism, whereas the T-A haplotype was associated with lower levels of neuroticism. Neuroticism has repeatedly been reported as a predictor and potential endophenotype for several psychiatric disorders, including major depression and schizophrenia (24 – 28) .
The observation that identical G72 haplotypes are not only associated with schizophrenia, bipolar disorder, panic disorder, and major depression but also with the personality dimension neuroticism suggests that G72 may confer susceptibility to major psychiatric disorders through trait anxiety, which is shared by different diagnostic entities. Although replication of our findings in other samples of different genetic background is clearly needed, we would like to argue the case for a psychiatric genetic research framework that complements disorder-focused genetic association testing by the study of easily measurable intermediate phenotypes, e.g., personality dimensions, in large samples from the general population. Such an approach may further close the sometimes existing gap between disorder-specific genetics and sophisticated neurobiological endophenotypic strategies (76) .

1. Chumakov I, Blumenfeld M, Guerassimenko O, Cavarec L, Palicio M, Abderrahim H, Bougueleret L, Barry C, Tanaka H, La Rosa P, Puech A, Tahri N, Cohen-Akenine A, Delabrosse S, Lissarrangue S, Picard FP, Maurice K, Essioux L, Millasseau P, Grel P, Debailleul V, Simon AM, Caterina D, Dufaure I, Malekzadeh K, Belova M, Luan JJ, Bouillot M, Sambucy JL, Primas G, Saumier M, Boubkiri N, Martin-Saumier S, Nasroune M, Peixoto H, Delaye A, Pinchot V, Bastucci M, Guillou S, Chevillon M, Sainz-Fuertes R, Meguenni S, Aurich-Costa J, Cherif D, Gimalac A, Van Duijn C, Gauvreau D, Ouellette G, Fortier I, Raelson J, Sherbatich T, Riazanskaia N, Rogaev E, Raeymaekers P, Aerssens J, Konings F, Luyten W, Macciardi F, Sham PC, Straub RE, Weinberger DR, Cohen N, Cohen D: Genetic and physiological data implicating the new human gene G72 and the gene for D-amino acid oxidase in schizophrenia. Proc Natl Acad Sci U S A 2002; 99:13675–13680; correction, 99:17221Google Scholar
2. Hattori E, Liu C, Badner JA, Bonner TI, Christian SL, Maheshwari M, Detera-Wadleigh SD, Gibbs RA, Gershon ES: Polymorphisms at the G72/G30 gene locus, on 13q33, are associated with bipolar disorder in two independent pedigree series. Am J Hum Genet 2003; 72:1131–1140Google Scholar
3. Chen YS, Akula N, Detera-Wadleigh SD, Schulze TG, Thomas J, Potash JB, DePaulo JR, McInnis MG, Cox NJ, McMahon FJ: Findings in an independent sample support an association between bipolar affective disorder and the G72/G30 locus on chromosome 13q33. Mol Psychiatry 2004; 9:87–92; correction, 9:811Google Scholar
4. Schumacher J, Jamra RA, Freudenberg J, Becker T, Ohlraun S, Otte AC, Tullius M, Kovalenko S, Bogaert AV, Maier W, Rietschel M, Propping P, Nöthen MM, Cichon S: Examination of G72 and D-amino-acid oxidase as genetic risk factors for schizophrenia and bipolar affective disorder. Mol Psychiatry 2004; 9:203–207Google Scholar
5. Detera-Wadleigh SD, McMahon FJ: G72/G30 in schizophrenia and bipolar disorder: review and meta-analysis. Biol Psychiatry 2006; 60:106–114Google Scholar
6. Fallin MD, Lasseter VK, Avramopoulos D, Nicodemus KK, Wolyniec PS, McGrath JA, Steel G, Nestadt G, Liang KY, Huganir RL, Valle D, Pulver AE: Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet 2005; 77:918–936Google Scholar
7. Ma J, Qin W, Wang XY, Guo TW, Bian L, Duan SW, Li XW, Zou FG, Fang YR, Fang JX, Feng GY, Gu NF, St Clair D, He L: Further evidence for the association between G72/G30 genes and schizophrenia in two ethnically distinct populations. Mol Psychiatry 2006; 11:479–487Google Scholar
8. Hong CJ, Hou SJ, Yen FC, Liou YJ, Tsai SJ: Family-based association study between G72/G30 genetic polymorphism and schizophrenia. Neuroreport 2006; 17:1067–1069Google Scholar
9. Liu YL, Fann CS, Liu CM, Chang CC, Wu JY, Hung SI, Liu SK, Hsieh MH, Hwang TJ, Chan HY, Chen JJ, Faraone SV, Tsuang MT, Chen WJ, Hwu HG: No association of G72 and D-amino acid oxidase genes with schizophrenia. Schizophr Res 2006; 87:15–20Google Scholar
10. Craddock N, O’Donovan MC, Owen MJ: The genetics of schizophrenia and bipolar disorder: dissecting psychosis. J Med Genet 2005; 42:193–204Google Scholar
11. Craddock N, Owen MJ: The beginning of the end for the Kraepelinian dichotomy. Br J Psychiatry 2005; 186:364–366Google Scholar
12. Green EK, Raybould R, Macgregor S, Gordon-Smith K, Heron J, Hyde S, Grozeva D, Hamshere M, Williams N, Owen MJ, O’Donovan MC, Jones L, Jones I, Kirov G, Craddock N: Operation of the schizophrenia susceptibility gene, neuregulin 1, across traditional diagnostic boundaries to increase risk for bipolar disorder. Arch Gen Psychiatry 2005; 62:642–648Google Scholar
13. Craddock N, O’Donovan MC, Owen MJ: Genes for schizophrenia and bipolar disorder? implications for psychiatric nosology. Schizophr Bull 2006; 32:9–16Google Scholar
14. Williams NM, Green EK, Macgregor S, Dwyer S, Norton N, Williams H, Raybould R, Grozeva D, Hamshere M, Zammit S, Jones L, Cardno A, Kirov G, Jones I, O’Donovan MC, Owen MJ, Craddock N: Variation at the DAOA/G30 locus influences susceptibility to major mood episodes but not psychosis in schizophrenia and bipolar disorder. Arch Gen Psychiatry 2006; 63:366–373Google Scholar
15. Maier W, Hofgen B, Zobel A, Rietschel M: Genetic models of schizophrenia and bipolar disorder: overlapping inheritance or discrete genotypes? Eur Arch Psychiatry Clin Neurosci 2005; 255:159–166Google Scholar
16. Abou Jamra R, Schmael C, Cichon S, Rietschel M, Schumacher J, Nöthen MM: The G72/G30 gene locus in psychiatric disorders: a challenge to diagnostic boundaries? Schizophr Bull 2006; 32:599–608Google Scholar
17. Ross CA, Margolis RL, Reading SA, Pletnikov M, Coyle JT: Neurobiology of schizophrenia. Neuron 2006; 52:139–153Google Scholar
18. Kato T: Molecular genetics of bipolar disorder and depression. Psychiatry Clin Neurosci 2007; 61:3–19Google Scholar
19. Schumacher J, Abou Jamra R, Becker T, Klopp N, Franke P, Jacob C, Sand P, Fritze J, Ohlraun S, Schulze TG, Rietschel M, Illig T, Propping P, Cichon S, Deckert J, Nöthen MM: Investigation of the DAOA/G30 locus in panic disorder. Mol Psychiatry 2005; 10:428–429Google Scholar
20. Gershon ES, DeLisi LE, Hamovit J, Nurnberger JI Jr, Maxwell ME, Schreiber J, Dauphinais D, Dingman CW II, Guroff JJ: A controlled family study of chronic psychoses: schizophrenia and schizoaffective disorder. Arch Gen Psychiatry 1988; 45:328–336Google Scholar
21. Alonso J, Angermeyer MC, Bernert S, Bruffaerts R, Brugha TS, Bryson H, de Girolamo G, Graaf R, Demyttenaere K, Gasquet I, Haro JM, Katz SJ, Kessler RC, Kovess V, Lépine JP, Ormel J, Polidori G, Russo LJ, Vilagut G, Almansa J, Arbabzadeh-Bouchez S, Autonell J, Bernal M, Buist-Bouwman MA, Codony M, Domingo-Salvany A, Ferrer M, Joo SS, Martínez-Alonso M, Matschinger H, Mazzi F, Morgan Z, Morosini P, Palacín C, Romera B, Taub N, Vollebergh WA, ESEMeD/MHEDEA 2000 Investigators, European Study of the Epidemiology of Mental Disorders (ESEMeD) Project: 12-Month comorbidity patterns and associated factors in Europe: results from the European Study of the Epidemiology of Mental Disorders (ESEMeD) project. Acta Psychiatr Scand Suppl 2004; 28–37Google Scholar
22. Kessler RC, Chiu WT, Demler O, Merikangas KR, Walters EE: Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry 2005; 62:617–627Google Scholar
23. McGuffin P, Knight J, Breen G, Brewster S, Boyd PR, Craddock N, Gill M, Korszun A, Maier W, Middleton T, Mors O, Owen MJ, Perry J, Preisig M, Reich T, Rice J, Rietschel M, Jones L, Sham P, Farmer AE: Whole genome linkage scan of recurrent depressive disorder from the depression network study. Hum Mol Genet 2005; 14:3337–3345Google Scholar
24. Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ: A longitudinal twin study of personality and major depression in women. Arch Gen Psychiatry 1993; 50:853–862Google Scholar
25. Duggan C, Sham P, Lee A, Minne C, Murray R: Neuroticism: a vulnerability marker for depression evidence from a family study. J Affect Disord 1995; 35:139–143Google Scholar
26. Kendler KS, Gatz M, Gardner CO, Pedersen NL: Personality and major depression: a Swedish longitudinal, population-based twin study. Arch Gen Psychiatry 2006; 63:1113–1120Google Scholar
27. Khan AA, Jacobson KC, Gardner CO, Prescott CA, Kendler KS: Personality and comorbidity of common psychiatric disorders. Br J Psychiatry 2005; 186:190–196Google Scholar
28. Van Os J, Jones PB: Neuroticism as a risk factor for schizophrenia. Psychol Med 2001; 31:1129–1134Google Scholar
29. Schulze TG, Ohlraun S, Czerski PM, Schumacher J, Kassem L, Deschner M, Gross M, Tullius M, Heidmann V, Kovalenko S, Jamra RA, Becker T, Leszczynska-Rodziewicz A, Hauser J, Illig T, Klopp N, Wellek S, Cichon S, Henn FA, McMahan FJ, Maier W, Propping P, Nöthen MM, Rietschel M: Genotype-phenotype studies in bipolar disorder showing association between the DAOA/G30 locus and persecutory delusions: a first step toward a molecular genetic classification of psychiatric phenotypes. Am J Psychiatry 2005; 162:2101–2108Google Scholar
30. Leckman JF, Sholomskas D, Thompson WD, Belanger A, Weissman MM: Best estimate of lifetime psychiatric diagnosis: a methodological study. Arch Gen Psychiatry 1982; 39:879–883Google Scholar
31. Spitzer RL, Williams JB, Gibbon M, First MB: The Structured Clinical Interview for DSM-III-R (SCID), I: history, rationale, and description. Arch Gen Psychiatry 1982; 49:624–629Google Scholar
32. Farmer AE, Wessely S, Castle D, McGuffin P: Methodological issues in using a polydiagnostic approach to define psychotic illness. Br J Psychiatry 1992; 161:824–830Google Scholar
33. Fangerau H, Ohlraun S, Granath RO, Nothen MM, Rietschel M, Schulze TG: Computer-assisted phenotype characterization for genetic research in psychiatry. Hum Hered 2004; 58:122–130Google Scholar
34. Hoefgen B, Schulze TG, Ohlraun S, von Widdern O, Höfels S, Gross M, Heidmann V, Kovalenko S, Eckermann A, Kölsch H, Metten M, Zobel A, Becker T, Nöthen MM, Propping P, Heun R, Maier W, Rietschel M: The power of sample size and homogenous sampling: association between the 5-HTTLPR serotonin transporter polymorphism and major depressive disorder. Biol Psychiatry 2005; 57:247–251Google Scholar
35. Costa PT, McCrae RR: Revised NEO Personality Inventory (NEO PI-R) and NEO Five Factor Inventory (NEO-FFI) Manual. Odessa, Psychological Assessment Resources, 1992Google Scholar
36. Barrett JC, Fry B, Maller J, Daly MJ: Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21:263–265Google Scholar
37. Dudbridge F: Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol 2003; 25:115–121Google Scholar
38. Beckmann L, Thomas DC, Fischer C, Chang-Claude J: Haplotype sharing analysis using Mantel statistics. Hum Hered 2005; 59:67–78Google Scholar
39. Te Meerman GJ, Van der Meulen MA, Sandkuijl LA: Perspectives of identity by descent (IBD) mapping in founder populations. Clin Exp Allergy 1995; 25:(suppl 2):97–102Google Scholar
40. Nordborg M, Innan H: Molecular population genetics. Curr Opin Plant Biol 2002; 5:69–73Google Scholar
41. Freimer NB, Heutink P, Wijmenga C: Obituary: Lodewijk A Sandkuijl, MD (July 31, 1953–Dec 4, 2002). Am J Hum Genet 2003; 72:781–784Google Scholar
42. Houwen RH, Baharloo S, Blankenship K, Raeymaekers P, Juyn J, Sandkuijl LA, Freimer NB: Genome screening by searching for shared segments: mapping a gene for benign recurrent intrahepatic cholestasis. Nat Genet 1994; 8:380–386Google Scholar
43. Van der Meulen MA, te Meerman GJ: Haplotype sharing analysis in affected individuals from nuclear families with at least one affected offspring. Genet Epidemiol 1997; 14:915–920Google Scholar
44. Dorum A, Abeler VM, Heimdal K, Trope C, Moller P: The problem of skipped generation and subclinical disease in familial breast-ovarian cancer. Acta Obstet Gynecol Scand 1997; 76:166–168Google Scholar
45. Boon M, Nolte IM, Bruinenberg M, Spijker GT, Terpstra P, Raelson J, De Keyser J, Zwanikken CP, Hulsbeek M, Hofstra RM, Buys CH, te Meerman GJ: Mapping of a susceptibility gene for multiple sclerosis to the 51 kb interval between G511525 and D6S1666 using a new method of haplotype sharing analysis. Neurogenetics 2001; 3:221–230Google Scholar
46. Levinson DF, Nolte I, Te Meerman GJ: Haplotype sharing tests of linkage disequilibrium in a Hutterite asthma data set. Genet Epidemiol 2001; 21(suppl 1):S308–S311Google Scholar
47. Sonneveld DJ, Lutke Holzik MF, Nolte IM, Sleijfer DT, van der Graaf WT, Bruinenberg M, Sijmons RH, Hoekstra HJ, Te Meerman GJ: Testicular carcinoma and HLA Class II genes. Cancer 2002; 95:1857–1863Google Scholar
48. Foerster J, Nolte I, Schweiger S, Ehlert C, Bruinenberg M, Spaar K, van der Steege G, Mulder M, Kalscheuer V, Moser B, Kijas Z, Seeman P, Ständer M, Sterry W, te Meerman G: Evaluation of the IRF-2 gene as a candidate for PSORS3. J Invest Dermatol 2004; 122:61–64Google Scholar
49. Beckmann L, Ziegler A, Duggal P, Bailey-Wilson JE: Haplotypes and haplotype-tagging single-nucleotide polymorphism: presentation group 8 of Genetic Analysis Workshop 14. Genet Epidemiol 2005; 29(suppl 1):S59–S71Google Scholar
50. Mantel N: The detection of disease clustering and a generalized regression approach. Cancer Res 1967; 27:209–220Google Scholar
51. Scheet P, Stephens M: A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet 2006; 78:629–644Google Scholar
52. Obreiter M, Fischer C, Chang-Claude J, Beckmann L: SDMinP: a program to control the family wise error rate using step-down minP adjusted P-values. Bioinformatics 2005; 21:3183–3184Google Scholar
53. Purcell S, Cherny SS, Sham PC: Genetic power calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 2003; 19:149–150Google Scholar
54. Dupont WD, Plummer WD: PS power and sample size program available for free on the Internet. Controlled Clin Trials 1997; 18:274Google Scholar
55. Hamilton SP, Fyer AJ, Durner M, Heiman GA, Baisre de Leon A, Hodge SE, Knowles JA, Weissman MM: Further genetic evidence for a panic disorder syndrome mapping to chromosome 13q. Proc Natl Acad Sci U S A 2003; 100:2550–2555Google Scholar
56. Li D, He L: G72/G30 genes and schizophrenia: a systematic meta-analysis of association studies. Genetics 2007; 175:917–922Google Scholar
57. Corvin A, McGhee KA, Murphy K, Donohoe G, Nangle JM, Schwaiger S, Kenny N, Clarke S, Meagher D, Quinn J, Scully P, Baldwin P, Browne D, Walsh C, Waddington JL, Morris DW, Gill M: Evidence for association and epistasis at the DAOA/G30 and D-amino acid oxidase loci in an Irish schizophrenia sample. Am J Med Genet B Neuropsychiatr Genet 2007; 144:949–953Google Scholar
58. Mutsuddi M, Morris DW, Waggoner SG, Daly MJ, Scolnick EM: Analysis of high-resolution hapmap of dtnbp1 (Dysbindin) suggests no consistency between reported common variant associations and schizophrenia. Am J Hum Genet 2006; 79:903–909Google Scholar
59. Lin PI, Vance JM, Pericak-Vance MA, Martin ER: No gene is an island: the flip-flop phenomenon. Am J Hum Genet 2007; 80:531–538Google Scholar
60. Kvajo M, Dhilla A, Swor DE, Karayiorgou M, Gogos JA: Evidence implicating the candidate schizophrenia/bipolar disorder susceptibility gene G72 in mitochondrial function. Mol Psychiatry 2007 (Epub ahead of print)Google Scholar
61. Schulze TG, McMahon FJ: Genetic linkage and association studies in bipolar affective disorder: a time for optimism. Am J Med Genet C Semin Med Genet 2003; 123:36–47Google Scholar
62. Comings DE: The real problem in association studies. Am J Med Genet B Neuropsychiatr Genet 2003; 116:102Google Scholar
63. Owen MJ: Genomic approaches to schizophrenia. Clin Ther 2005; 27(suppl A):S2–S7Google Scholar
64. Johnson C, Drgon T, Liu QR, Walther D, Edenberg H, Rice J, Foroud T, Uhl GR: Pooled association genome scanning for alcohol dependence using 104,268 SNPs: validation and use to identify alcoholism vulnerability loci in unrelated individuals from the collaborative study on the genetics of alcoholism. Am J Med Genet B Neuropsychiatr Genet 2006; 141:844–853Google Scholar
65. Keller MC, Miller G: Resolving the paradox of common, harmful, heritable mental disorders: which evolutionary genetic models work best? Behav Brain Sci 2006; 29:385–404Google Scholar
66. Smyth DJ, Cooper JD, Bailey R, Field S, Burren O, Smink LJ, Guja C, Ionescu-Tirgoviste C, Widmer B, Dunger DB, Savage DA, Walker NM, Clayton DG, Todd JA: A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet 2006; 38:617–619Google Scholar
67. Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P: A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007; 445:881–885Google Scholar
68. Baum AE, Akula N, Cabanero M, Cardona I, Corona W, Klemens B, Schulze TG, Cichon S, Rietschel M, Nöthen MM, Georgi A, Schumacher J, Schwarz M, Abou Jamra R, Höfels S, Propping P, Satagopan J, Detera-Wadleigh SD, Hardy J, McMahon FJ: A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry 2008; 13:197–207Google Scholar
69. Wellcome Trust Case Control Consortium: Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007; 447:661–678Google Scholar
70. Kendler KS, Eaves LJ, Walters EE, Neale MC, Heath AC, Kessler RC: The identification and validation of distinct depressive syndromes in a population-based sample of female twins. Arch Gen Psychiatry 1996; 53:391–399Google Scholar
71. Camp NJ, Cannon-Albright LA: Dissecting the genetic etiology of major depressive disorder using linkage analysis. Trends Mol Med 2005; 11:138–144Google Scholar
72. Gottesman II, Gould TD: The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry 2003; 160:636–645Google Scholar
73. Schulze TG, McMahon FJ: Defining the phenotype in human genetic studies: forward genetics and reverse phenotyping. Hum Hered 2004; 58:131–138Google Scholar
74. Freeman D, Garety PA: Comments on the content of persecutory delusions: does the definition need clarification? Br J Clin Psychol 2000; 39:407–414Google Scholar
75. Startup H, Freeman D, Garety PA: Persecutory delusions and catastrophic worry in psychosis: developing the understanding of delusion distress and persistence. Behav Res Ther 2007; 45:523–537Google Scholar
76. Walters JTR, Owen MJ: Endophenotypes in psychiatric genetics. Mol Psychiatry 2007; 12:886–890Google Scholar