Suggestive Linkage to Chromosomal Regions 13q31 and 22q12 in Families With Psychotic Bipolar Disorder
Abstract
OBJECTIVE: Linkage studies of bipolar disorder and schizophrenia have found overlapping evidence for susceptibility genes in four chromosomal regions—10p12-14, 13q32, 18p11.2, and 22q12-13. The authors previously demonstrated familial clustering of psychotic symptoms—defined as hallucinations and/or delusions—in some bipolar disorder pedigrees. In this study they used stratified linkage analysis to test the hypothesis that those bipolar disorder pedigrees most enriched for psychotic symptoms would show greater evidence of linkage to the regions of previous bipolar disorder/schizophrenia linkage overlap. METHOD: Nonparametric linkage analyses using GENEHUNTER and ASPEX were performed on 65 bipolar disorder families. Family subsets were defined by the number of family members with psychotic mood disorder. RESULTS: The 10 families in which three or more members had psychotic mood disorder showed suggestive evidence of linkage to 13q31 (nonparametric linkage score=3.56; LOD score=2.52) and 22q12 (nonparametric linkage score=3.32; LOD score=3.06). These results differed significantly from those for the entire study group of 65 families, which showed little or no linkage evidence in the two regions. The 10 families with three or more psychotic members did not show evidence of linkage to 10p12-14 or 18p11.2. The 95% confidence interval on 22q12 spanned 4.3 centimorgans (2.6 megabases) and was congruent with previous findings. CONCLUSIONS: Bipolar disorder families in which psychotic symptoms cluster may carry susceptibility genes on chromosomal regions 13q31 and 22q12. Replication should be attempted in similar families and perhaps in schizophrenia families in which mood symptoms cluster because these overlapping phenotypes may correlate most closely with the putative susceptibility genes. The localization of the 22q12 finding particularly encourages further study of this region.
Overlap between bipolar disorder and schizophrenia has long been noted clinically. About 9% of people with schizophrenia have manic syndromes (1) and 25% have depressive syndromes (2); psychotic symptoms—hallucinations and delusions—occur in 58% of people with bipolar I disorder (3). The existence of a diagnostic category incorporating elements of mood disorder and schizophrenia—schizoaffective disorder—reflects the difficulty that occasionally arises in distinguishing the two illnesses.
Evidence for overlapping heritability of bipolar disorder and schizophrenia has emerged from genetic epidemiologic studies. Family studies have provided the strongest evidence. An excess of major depression has been observed in relatives of probands with bipolar disorder and relatives of probands with schizophrenia (see reference 4 for references). Most studies, however, have not found an excess of relatives with schizophrenia in families of probands with bipolar disorder or an excess of relatives with bipolar disorder in families of schizophrenia probands. Studies from three data sets have addressed the question of overlap between psychotic mood disorder and schizophrenia. Two found elevated rates of psychotic mood disorder in relatives of probands with schizophrenia and vice versa; the third also suggested shared liability between the two entities (see reference 4 for references). In addition, some twin studies found evidence of shared heritability between psychotic mood disorder and schizophrenia (1, 5).
Linkage studies in bipolar disorder and schizophrenia have implicated four overlapping chromosomal regions that could harbor susceptibility genes shared by the two disorders (6). On chromosome 13q32, two significant findings (7, 8) and a more modest one (9) were reported in schizophrenia, although one large replication attempt was negative (10). Two suggestive findings were reported in bipolar disorder (11, 12). On chromosome 22q12-13, modest evidence for linkage in schizophrenia was reported (13, 14); significant linkage for bipolar disorder was found in the same region (12). A recent meta-analysis of 11 bipolar disorder and 18 schizophrenia genome scans found that 13q32 and 22q12 were the two most significantly linked chromosomal regions for each of the two disorders (15). On chromosome 10p12-14, three studies found suggestive evidence for linkage in schizophrenia (16–18); one study found suggestive evidence in bipolar disorder (19). On chromosome 18p11.2, three studies found evidence for linkage in bipolar disorder (20–22) and one study implicated schizophrenia (23).
We have previously used clinical features to attempt to reduce genetic heterogeneity in bipolar disorder families. We found that bipolar II disorder enhanced evidence for linkage to chromosome 18q (24). We also found that psychotic symptoms were familial in 65 bipolar disorder pedigrees (25). The current study used psychotic symptoms to attempt to reduce genetic heterogeneity in these families. We reasoned that bipolar disorder families in which psychotic symptoms were prominent would be most likely to carry susceptibility genes shared with schizophrenia. These same families would thus be more likely to show linkage to the four chromosomal regions of bipolar disorder/schizophrenia linkage overlap than would other bipolar disorder families. Linkage in these regions was assessed for all families and for three family subsets defined by the number of family members with psychotic mood disorder.
Method
Subjects
Probands were selected from patients screened in Baltimore and Iowa City and from volunteer patients who contacted us. The ascertainment criteria required a treated bipolar I disorder proband and at least two first-degree relatives with a major mood disorder—bipolar I disorder, bipolar II disorder, recurrent unipolar depression, or schizoaffective disorder, manic type. Probands and relatives were interviewed by psychiatrists using the Schedule for Affective Disorders and Schizophrenia—Lifetime Version (26). After complete description of the study to subjects, written informed consent was obtained. Information from family informants was obtained, and data from medical records were added whenever possible. Mood disorder diagnoses were made by two independent psychiatrists using a best estimate procedure and Research Diagnostic Criteria (27).
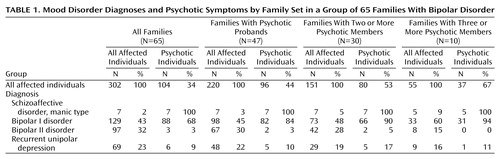
There were 65 bipolar I disorder probands and 237 affected relatives, including 64 relatives with bipolar I disorder, seven with schizoaffective disorder, manic type, 97 with bipolar II disorder, and 69 with recurrent unipolar depression. No subjects had schizophrenia or schizoaffective disorder, depressed type. A total of 104 subjects were psychotic as defined by presence of hallucinations and/or delusions during a mood episode. One additional newly ascertained psychotic bipolar I disorder subject had genotypes available only on chromosome 22 and was included for analyses of those markers. All subjects with a mood disorder were considered affected for linkage analysis. This single affection status was used because family studies suggest that major depression is genetically related both to bipolar disorder and to schizophrenia, and this study was designed to use phenotypic overlap of the two disorders to clarify potential genetic overlap. Stratification was performed on the basis of the presence of psychotic symptoms in subjects with mood disorder. Family groupings included 1) all families (65 families, 489 affected relative pairs, 325 affected sibling pairs); 2) families with psychotic probands (47 families, 373 affected relative pairs, 249 affected sibling pairs); 3) families with a psychotic proband and one or more other psychotic members (30 families, 270 affected relative pairs, 179 affected sibling pairs); and 4) families with three or more psychotic members (10 families, 116 affected relative pairs, 88 affected sibling pairs). Diagnostic breakdown and psychotic symptoms by family set are shown in Table 1.
Genotyping
Many of the genotypes used in this study were generated for a genome scan. Full description of the genotyping methods employed in this scan is provided in another publication (28). The markers consisted of microsatellite tandem repeats. Genotyping was done at Johns Hopkins, Stanford, and the Center for Inherited Disease Research. Additional markers were genotyped on chromosomes 13q31-32 and 22q12 to follow up positive preliminary results. Mendelian inconsistencies were resolved before linkage analysis by using the GAS program (http://users.ox.ac.uk/~ayoung/gas.html) and the UNKNOWN program from the LINKAGE package (ftp://linkage.rockefeller.edu/software/linkage/). The ASPEX SIBCLEAN program (ftp://lahmed.stanford.edu/pub/aspex) was used to check for double recombinants. Initial marker order was assessed by using the Marshfield map (http://research.marshfieldclinic.org/genetics/). We used CRI-MAP (http://compgen.rutgers.edu/multimap/crimap/index.html) to refine marker order further, taking account of physical data in the University of California, Santa Cruz, human genome browser, December 2001 assembly (http://www.genome.ucsc.edu). Average spacing of DNA markers was as follows: chromosome 10, 4 centimorgans (cM); chromosome 13, 4 cM; chromosome 18, 6 cM; and chromosome 22, 2 cM. Genetic information content is given in Figure 1.
Linkage Analysis
Nonparametric linkage analysis was performed with GENEHUNTER (version 2.0) with the ALL option (http://www.genome. wi.mit.edu/ftp/distribution/software/genehunter/), which tests identity-by-descent allele sharing among all affected individuals in each pedigree. To confirm results from GENEHUNTER, analyses were also performed with the ASPEX SIBIBD program (version 2.2) (ftp://lahmed.stanford.edu/pub/aspex). This program calculates maximum nonparametric LOD scores in affected sibling pairs. The nonparametric linkage score and the ASPEX LOD score are two among many linkage statistics. Such statistics are not equivalent in their absolute values; comparisons between them can be made only after conversion to the common currency of the p value.
The GENEFINDER program (http://biosun01.biostat.jhsph.edu/~wmchen/gf.html) was used to localize findings on chromosomes 13 and 22. This program uses the maximum likelihood estimate of identity-by-descent sharing from GENEHUNTER to calculate the most likely location of a disease gene and a 95% confidence interval for the location (29). This method can provide valuable information to complement the results of GENEHUNTER because the peak nonparametric linkage score does not necessarily correspond to the most likely site of the disease locus. A shifted peak can occur because more polymorphic markers may give rise to larger nonparametric linkage scores despite being further away from the disease locus than closer, less informative markers.
For GENEHUNTER findings on chromosomes 13 and 22, the empirical significance of the nonparametric linkage scores for the 10 families with three or more psychotic members compared with the entire group of families was assessed through simulation using the method of Cox et al. (30). Ten thousand samples of 10 families randomly selected from the total data set were generated, and nonparametric linkage scores for each sample were derived. A similar procedure was used to assess the empirical significance of the SIBIBD results.
Results
Results of the GENEHUNTER analysis are shown in Figure 2. The 10 families in which three or more members had psychotic mood disorder showed nominally significant evidence of linkage to 13q31 (nonparametric linkage score=3.56, p=0.003) at D13S317 and to 22q12 (nonparametric linkage score=3.32; p=0.005) at D22S277. At these peaks on both chromosomes 13 and 22 the evidence for linkage was successively lower as the number of psychotic subjects in families decreased. In the family set in which the proband and at least one other member had psychotic mood disorder, the nonparametric linkage score on 13q31 fell to 1.29 and the nonparametric linkage score on 22q12 fell to 2.30. In the psychotic proband family set, the scores dropped to –0.18 on 13q31 and 1.55 on 22q12. In the full family set, the scores were –0.28 on 13q31 and 0.63 on 22q12.
GENEHUNTER findings were negative on chromosomes 10 and 18. For both chromosomes no psychotic family set yielded nonparametric linkage scores in the significant or suggestive range. Further, in no psychotic family set did the nonparametric linkage scores differ significantly from those in the full family set.
Analyses on chromosomes 13 and 22 with SIBIBD in the 10 families with the most psychotic members yielded results similar to those found by using GENEHUNTER. For these 10 families, the maximum LOD score on 13q31 was 2.52 at D13S317 (p=0.0006) and the maximum LOD score on 22q12 was 3.06 (p=0.00009) at D22S277.
GENEFINDER was used to compute the most likely location of the putative susceptibility genes on chromosomes 13 and 22 and a 95% confidence interval for these locations. On chromosome 13 the estimated gene location was at 62.8 cM on the Marshfield map, 1.1 cM centromeric to D13S317, and the 95% confidence interval extended from 56.4 to 69.3 cM, a span of 12.9 cM. On chromosome 22 the estimated gene location was at 29.4 cM on the Marshfield map, 0.75 cM centromeric to D22S277, with the 95% confidence interval extending from 27.3 to 31.6 cM, a span of 4.3 cM.
Two simulations were conducted to determine empirical p values for the stratification of the families with the largest number of psychotic members. On 13q31, a nonparametric linkage score of ≥3.56 was observed for randomly selected sets of 10 families 22 times in 10,000 simulations—equivalent to an empirical p value of 0.002. On 22q12, a nonparametric linkage score of ≥3.32 was observed 242 times in 10,000 simulations—equivalent to an empirical p value of 0.024. Using SIBIBD, a 10,000-sample simulation yielded p values of 0.007 on 13q31 and 0.012 on 22q12.
Discussion
We examined four chromosomal regions with preexisting evidence for linkage to both bipolar disorder and schizophrenia. In two of the four regions, 13q31 and 22q12, the families with the highest number of psychotic members showed suggestive evidence of linkage, and a “dose-response”-like effect was observed between the number of family members with psychotic symptoms and the strength of linkage. Linkage evidence obtained by using one algorithm was supported by similar evidence obtained by using a different algorithm. Two simulations found that the results on 13q31 and 22q12 in the 10 families with the most psychotic members differed significantly from that expected by chance in this 65-family data set, suggesting that stratification by number of family members with psychotic mood disorder may be genetically meaningful.
Relation to Previous Findings
The linkage peak on 22q12 near D22S277 is congruent with findings from previous studies. The only previous significant result in bipolar disorder found linkage at D22S278 (12). A meta-analysis of schizophrenia studies also found modest support for linkage at D22S278 (14). D22S277 and D22S278 are just 0.135 megabases (Mb) apart physically and are indistinguishable on a genetic recombination map. In a meta-analysis of 29 genome scans of bipolar disorder and schizophrenia (15), 22q12 showed the strongest linkage evidence of any chromosomal region (p=0.00000002). This linkage region is about 16 Mb physically from the velocardiofacial syndrome region of chromosome 22q11.2.
On 13q31 the peak nonparametric linkage score occurred 15–29 cM centromeric to those identified by previous linkage studies (7, 8, 11, 12), and 28 cM centromeric to the G72 gene that was recently implicated in schizophrenia susceptibility (31). Although not entirely congruent with previous findings, our result may represent the effects of the same disease locus as the previous reports because of the low resolution of linkage studies (32). With 200 or fewer study families, a replication of a previous true finding may show a signal up to 30 cM away from the original finding. The 29-genome scan meta-analysis (15) found that 13q32 was the second most strongly linked chromosomal region (p=0.0000002).
Etiologic Overlap With Schizophrenia
The present findings support the hypothesis that psychotic symptoms define a subtype of bipolar disorder with etiologic distinction from broader bipolar disorder and etiologic overlap with schizophrenia. The etiologic overlap hypothesis suggests the existence of either psychosis genes or joint mood/psychosis genes. This latter possibility would imply that examination of mood-related subtypes of schizophrenia would further illuminate the issue of etiologic overlap. Indeed, two studies found some evidence that elevated familial rates of mood disorder occur in a subset of subjects with schizophrenia (33, 34). The hypothesis that some families with schizophrenia might carry joint mood/psychosis genes was tested by Pulver et al. (35). To try to reduce genetic heterogeneity in schizophrenia, they analyzed a subset of six families (from a set of 54) containing at least one relative with psychotic mood disorder. The strongest linkage signal in the genome for this subset was on chromosome 22q12; results were suggestive. Linkage evidence was also suggestive for 13q33.
Localization of Putative Susceptibility Gene
Although the present finding on 22q12 is only suggestive rather than significant, the narrow 95% confidence interval, the congruence with previous findings, and the weight of the meta-analytic linkage evidence encourage pursuit of a susceptibility gene in this region. The 95% confidence interval of 4.3 cM likely represents 2.6 Mb of physical distance (http://www.genome.ucsc.edu). On chromosome 13, our finding is less clearly localized. The 95% confidence interval of 12.9 cM likely represents 21.1 Mb. Our finding on chromosome 13 is also 15–29 cM centromeric to previous findings, which introduces further uncertainty concerning localization.
Given its completed sequencing, chromosome 22 presents an especially attractive target for gene-finding. The advanced state of gene characterization and single nucleotide polymorphism identification should facilitate the search for bipolar disorder/schizophrenia genes. In this 2.6 Mb interval there are 25 annotated genes, 16 of which have been characterized. Of particular interest is apolipoprotein L2, which has been shown to be up-regulated in postmortem schizophrenia and bipolar disorder brains (36). Single nucleotide polymorphism genotyping across this region to test for linkage disequilibrium may help to narrow the localization further.
Methodologic Considerations
There are several strengths in the methodology of this study. First, clinical assessment was rigorously conducted. The best estimate procedure was used to assign diagnoses with high reliability. The assessment of psychotic symptoms was performed on the basis of both interview data and supplementary data from family informants as well as data from medical records. Second, follow-up genotyping ensured good information content for 13q31-32 and 22q12. Third, the analysis was hypothesis-driven in that the four chromosomal regions selected demonstrated previous evidence of linkage overlap between bipolar disorder and schizophrenia.
This study should also be viewed in light of several limitations. First, alternative ways to define psychotic bipolar disorder families could be construed. Even in the 10 families with the most psychotic members, not all subjects with mood disorder had psychotic symptoms. There were only five families, however, in which all subjects with mood disorders were psychotic, so the power to detect linkage in this subset would have been low.
Second, the nonpsychotic subjects with mood disorders in the 10 families with the most psychotic members contributed to the evidence for linkage on chromosomes 13q and 22q. We speculate that those nonpsychotic subjects could be 13q/22q susceptibility allele carriers who fail to express psychotic symptoms because they lack susceptibility alleles for other interacting genes, or because they lack exposure to interacting environmental factors.
Third, clinical variables that could co-occur with psychosis, such as illness chronicity or cognitive impairment, might be more genetically relevant than psychosis itself. Although our data did not allow adequate assessment of these variables, we did compare the 10 families with the most psychotic members and the 55 other families on age at onset of mood disorder, number of mood disorder episodes, and duration of longest mood disorder episode; no significant differences were found (data not shown). Additionally, the two family sets did not differ significantly in rates of panic disorder or panic attack, clinical features associated with linkage to chromosome 18q22 (data not shown).
Fourth, the absence of linkage signals on chromosomes 10p and 18p could represent false negative findings because the power in the 10 families with the most psychotic members to detect genes of small effect was low and because, on chromosome 10p in particular, the genetic information content was modest.
Fifth, the statistical significance of these findings is difficult to assess in the light of the widely used Lander and Kruglyak criteria (37). These findings might be considered “confirmed” linkage because this study attempted replication of four regions previously identified by bipolar disorder and schizophrenia linkage studies. However, the more conservative interpretation of “suggestive” linkage might be more appropriate because this report used a novel phenotypic family characterization rather than simple bipolar disorder.
In summary, we found that 10 families with psychotic bipolar disorder demonstrate suggestive linkage to two chromosomal regions, 13q31 and 22q12, which are in or near regions previously implicated in both bipolar disorder and schizophrenia. Replication should be attempted in other bipolar disorder families in which psychotic symptoms cluster and, perhaps, in schizophrenia families in which mood symptoms cluster because these overlapping phenotypes may be most closely associated with the putative susceptibility loci. The localization of the 22q12 finding particularly encourages further study of this region.
 |
Presented in part at the American Society for Human Genetics 50th Annual Meeting, Philadelphia, Oct. 3–7, 2000, and the Xth World Congress of Psychiatric Genetics, Brussels, Oct. 9–13, 2002. Received Aug. 16, 2002; revision received Dec. 13, 2002; accepted Jan. 22, 2003. From the Department of Psychiatry and Behavioral Sciences, Johns Hopkins School of Medicine, Baltimore; the Departments of Epidemiology and Mental Hygiene, Johns Hopkins Bloomberg School of Public Health, Baltimore; the Department of Psychiatry, Yu-Li Veterans Hospital, Hualien, Taiwan; the Department of Psychiatry, University of Colorado School of Medicine, Denver; and the Department of Psychiatry, University of Chicago. Address reprint requests to Dr. Potash, Meyer 3-181, Johns Hopkins Hospital, 600 N. Wolfe St., Baltimore, MD 21287-7381; [email protected] (e-mail). Supported by NIMH research grants MH-42243 (Dr. DePaulo) and MH-02026 (Dr. Potash), the National Alliance for Research on Schizophrenia and Depression, the Stanley Medical Research Institute, the Dana Foundation, the Alexander Wilson Schweizer Fund, the Affective Disorders Fund, and the George Browne Laboratory Fund. The authors thank Koustubh Ranade, Carl Friddle, David Botstein, O. Colin Stine, and the Center for Inherited Disease Research for genotyping data; Rebecca Koskella, Thomas Marr, Theresa Swift-Scanlan, Jen Chellis, Amy Mahoney, and Barbara Schweizer for data management; and Thomas Schulze, Ann Pulver, and Andrew Feinberg for critical input.
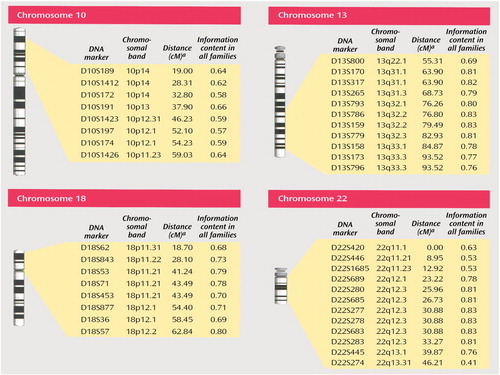
Figure 1. Location and Information Content of DNA Markers Used for Genotyping
aFrom the Marshfield map (http://research.marshfieldclinic.org/genetics/).
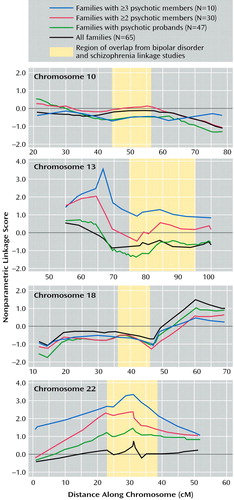
Figure 2. Nonparametric Linkage Scores in Bipolar Disorder Families, Stratified by Presence of Psychotic Symptoms in Affected Family Members
1. Cardno AG, Rijsdijk FV, Sham PC, Murray RM, McGuffin P: A twin study of genetic relationships between psychotic symptoms. Am J Psychiatry 2002; 159:539-545Link, Google Scholar
2. Siris SG: Depression in schizophrenia: perspective in the era of “atypical” antipsychotic agents. Am J Psychiatry 2000; 157:1379-1389Link, Google Scholar
3. Goodwin FK, Jamison KR: Manic-Depressive Illness. New York, Oxford University Press, 1990Google Scholar
4. Potash JB, Chiu YF, MacKinnon DF, Miller EB, Simpson SG, McMahon FJ, McInnis MG, DePaulo JR Jr: Familial aggregation of psychotic symptoms in a replication set of 69 bipolar disorder pedigrees. Am J Med Genet 2003; 116B:90-97Google Scholar
5. Farmer AE, McGuffin P, Gottesman II: Twin concordance for DSM-III schizophrenia: scrutinizing the validity of the definition. Arch Gen Psychiatry 1987; 44:634-641Crossref, Medline, Google Scholar
6. Berrettini WH: Are schizophrenic and bipolar disorders related? a review of family and molecular studies. Biol Psychiatry 2000; 48:531-538Crossref, Medline, Google Scholar
7. Blouin JL, Dombroski BA, Nath SK, Lasseter VK, Wolyniec PS, Nestadt G, Thornquist M, Ullrich G, McGrath J, Kasch L, Lamacz M, Thomas MG, Gehrig C, Radhakrishna U, Snyder SE, Balk KG, Neufeld K, Swartz KL, DeMarchi N, Papadimitriou GN, Dikeos DG, Stefanis CN, Chakravarti A, Childs B, Pulver AE: Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21. Nat Genet 1998; 20:70-73Crossref, Medline, Google Scholar
8. Brzustowicz LM, Honer WG, Chow EW, Little D, Hogan J, Hodgkinson K, Bassett AS: Linkage of familial schizophrenia to chromosome 13q32. Am J Hum Genet 1999; 65:1096-1103Crossref, Medline, Google Scholar
9. Lin MW, Sham P, Hwu HG, Collier D, Murray R, Powell JF: Suggestive evidence for linkage of schizophrenia to markers on chromosome 13 in Caucasian but not Oriental populations. Hum Genet 1997; 99:417-420Crossref, Medline, Google Scholar
10. Levinson DF, Holmans P, Straub RE, Owen MJ, Wildenauer DB, Gejman PV, Pulver AE, Laurent C, Kendler KS, Walsh D, Norton N, Williams NM, Schwab SG, Lerer B, Mowry BJ, Sanders AR, Antonarakis SE, Blouin JL, DeLeuze JF, Mallet J: Multicenter linkage study of schizophrenia candidate regions on chromosomes 5q, 6q, 10p, and 13q: schizophrenia linkage collaborative group III. Am J Hum Genet 2000; 67:652-663Crossref, Medline, Google Scholar
11. Detera-Wadleigh SD, Badner JA, Berrettini WH, Yoshikawa T, Goldin LR, Turner G, Rollins DY, Moses T, Sanders AR, Karkera JD, Esterling LE, Zeng J, Ferraro TN, Guroff JJ, Kazuba D, Maxwell ME, Nurnberger JIJ, Gershon ES: A high-density genome scan detects evidence for a bipolar-disorder susceptibility locus on 13q32 and other potential loci on 1q32 and 18p11.2. Proc Natl Acad Sci USA 1999; 96:5604-5609Crossref, Medline, Google Scholar
12. Kelsoe JR, Spence MA, Loetscher E, Foguet M, Sadovnick AD, Remick RA, Flodman P, Khristich J, Mroczkowski-Parker Z, Brown JL, Masser D, Ungerleider S, Rapaport MH, Wishart WL, Luebbert H: A genome survey indicates a possible susceptibility locus for bipolar disorder on chromosome 22. Proc Natl Acad Sci USA 2001; 98:585-590Crossref, Medline, Google Scholar
13. Pulver AE, Karayiorgou M, Wolyniec PS, Lasseter VK, Kasch L, Nestadt G, Antonarakis S, Housman D, Kazazian HH, Meyers D: Sequential strategy to identify a susceptibility gene for schizophrenia: report of potential linkage on chromosome 22q12-q13.1, part 1. Am J Med Genet 1994; 54:36-43Crossref, Medline, Google Scholar
14. Gill M, Vallada H, Collier D, Sham P, Holmans P, Murray R, McGuffin P, Nanko S, Owen M, Antonarakis S, Housman D, Kazazian H, Nestadt G, Pulver AE, Straub RE, MacLean CJ, Walsh D, Kendler KS, DeLisi L, Polymeropoulos M, Coon H, Byerley W, Lofthouse R, Gershon E, Read CM (Schizophrenia Collaborative Linkage Group [Chromosome 22]): A combined analysis of D22S278 marker alleles in affected sib-pairs: support for a susceptibility locus for schizophrenia at chromosome 22q12. Am J Med Genet 1996; 67:40-45Crossref, Medline, Google Scholar
15. Badner JA, Gershon ES: Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry 2002; 7:405-411Crossref, Medline, Google Scholar
16. Schwab SG, Hallmayer J, Albus M, Lerer B, Eckstein GN, Borrmann M, Segman RH, Hanses C, Freymann J, Yakir A, Trixler M, Falkai P, Rietschel M, Maier W, Wildenauer DB: A genome-wide autosomal screen for schizophrenia susceptibility loci in 71 families with affected siblings: support for loci on chromosome 10p and 6. Mol Psychiatry 2000; 5:638-649Crossref, Medline, Google Scholar
17. Straub RE, MacLean CJ, Martin RB, Ma Y, Myakishev MV, Harris-Kerr C, Webb BT, O’Neill FA, Walsh D, Kendler KS: A schizophrenia locus may be located in region 10p15-p11. Am J Med Genet 1998; 81:296-301Crossref, Medline, Google Scholar
18. Faraone SV, Matise T, Svrakic D, Pepple J, Malaspina D, Suarez B, Hampe C, Zambuto CT, Schmitt K, Meyer J, Markel P, Lee H, Harkavy FJ, Kaufmann C, Cloninger CR, Tsuang MT: Genome scan of European-American schizophrenia pedigrees: results of the NIMH Genetics Initiative and Millennium Consortium. Am J Med Genet 1998; 81:290-295Crossref, Medline, Google Scholar
19. Foroud T, Castelluccio PF, Koller DL, Edenberg HJ, Miller M, Bowman E, Rau NL, Smiley C, Rice JP, Goate A, Armstrong C, Bierut LJ, Reich T, Detera-Wadleigh SD, Goldin LR, Badner JA, Guroff JJ, Gershon ES, McMahon FJ, Simpson S, MacKinnon D, McInnis M, Stine OC, DePaulo JR, Blehar MC, Nurnberger JIJ: Suggestive evidence of a locus on chromosome 10p using the NIMH genetics initiative bipolar affective disorder pedigrees. Am J Med Genet 2000; 96:18-23Crossref, Medline, Google Scholar
20. Berrettini WH, Ferraro TN, Goldin LR, Weeks DE, Detera-Wadleigh S, Nurnberger JIJ, Gershon ES: Chromosome 18 DNA markers and manic-depressive illness: evidence for a susceptibility gene. Proc Natl Acad Sci USA 1994; 91:5918-5921Crossref, Medline, Google Scholar
21. Stine OC, Xu J, Koskela R, McMahon FJ, Gschwend M, Friddle C, Clark CD, McInnis MG, Simpson SG, Breschel TS, Vishio E, Riskin K, Feilotter H, Chen E, Folstein SE, Meyers DA, Botstein D, Marr TG, DePaulo JR: Evidence for linkage of bipolar disorder to chromosome 18 with a parent-of-origin effect. Am J Hum Genet 1995; 57:1384-1394Medline, Google Scholar
22. Nothen MM, Cichon S, Rohleder H, Hemmer S, Franzek E, Fritze J, Albus M, Borrmann-Hassenbach M, Kreiner R, Weigelt B, Minges J, Lichtermann D, Maier W, Craddock N, Fimmers R, Holler T, Baur MP, Rietschel M, Propping P: Evaluation of linkage of bipolar affective disorder to chromosome 18 in a sample of 57 German families. Mol Psychiatry 1999; 4:76-84Crossref, Medline, Google Scholar
23. Schwab SG, Hallmayer J, Lerer B, Albus M, Borrmann M, Honig S, Strauss M, Segman R, Lichtermann D, Knapp M, Trixler M, Maier W, Wildenauer DB: Support for a chromosome 18p locus conferring susceptibility to functional psychoses in families with schizophrenia, by association and linkage analysis. Am J Hum Genet 1998; 63:1139-1152Crossref, Medline, Google Scholar
24. McMahon FJ, Simpson SG, McInnis MG, Badner JA, MacKinnon DF, DePaulo JR: Linkage of bipolar disorder to chromosome 18q and the validity of bipolar II disorder. Arch Gen Psychiatry 2001; 58:1025-1031Crossref, Medline, Google Scholar
25. Potash JB, Willour VL, Chiu YF, Simpson SG, MacKinnon DF, Pearlson GD, DePaulo JR Jr, McInnis MG: The familial aggregation of psychotic symptoms in bipolar disorder pedigrees. Am J Psychiatry 2001; 158:1258-1264Link, Google Scholar
26. Endicott J, Spitzer RL: A diagnostic interview: the Schedule for Affective Disorders and Schizophrenia. Arch Gen Psychiatry 1978; 35:837-844Crossref, Medline, Google Scholar
27. Spitzer RL, Endicott J, Robins E: Research Diagnostic Criteria (RDC) for a Selected Group of Functional Disorders, 2nd ed. New York, New York State Psychiatric Institute, Biometrics Research, 1975Google Scholar
28. McInnis MG, Lan TH, Willour VL, McMahon FJ, Simpson SG, Addington AM, MacKinnon DF, Potash JB, Mahoney AT, Chellis J, Huo Y, Swift-Scanlan T, Chen H, Koskela R, Stine OC, Jamison KR, Holmans P, Folstein SE, Ranade K, Friddle C, Botstein D, Marr T, Beaty TH, Zandi P, DePaulo JR: Genome-wide scan of bipolar disorder in 65 pedigrees: supportive evidence for linkage at 8q24, 18q22, 4q32, 2p12, and 13q12. Mol Psychiatry (in press)Google Scholar
29. Liang KY, Chiu YF, Beaty TH: A robust identity-by-descent procedure using affected sib pairs: multipoint mapping for complex diseases. Hum Hered 2001; 51:64-78Crossref, Medline, Google Scholar
30. Cox NJ, Frigge M, Nicolae DL, Concannon P, Hanis CL, Bell GI, Kong A: Loci on chromosomes 2 (NIDDM1) and 15 interact to increase susceptibility to diabetes in Mexican Americans. Nat Genet 1999; 21:213-215Crossref, Medline, Google Scholar
31. Chumakov I, Blumenfeld M, Guerassimenko O, Cavarec L, Palicio M, Abderrahim H, Bougueleret L, Barry C, Tanaka H, La Rosa P, Puech A, Tahri N, et al: Genetic and physiological data implicating the new human gene G72 and the gene for D-amino acid oxidase in schizophrenia. Proc Natl Acad Sci USA 2002; 99:13675-13680; correction, 99:17221Crossref, Medline, Google Scholar
32. Roberts SB, MacLean CJ, Neale MC, Eaves LJ, Kendler KS: Replication of linkage studies of complex traits: an examination of variation in location estimates. Am J Hum Genet 1999; 65:876-884Crossref, Medline, Google Scholar
33. Sham PC, MacLean CJ, Kendler KS: A typological model of schizophrenia based on age at onset, sex and familial morbidity. Acta Psychiatr Scand 1994; 89:135-141Crossref, Medline, Google Scholar
34. Sham PC, Castle DJ, Wessely S, Farmer AE, Murray RM: Further exploration of a latent class typology of schizophrenia. Schizophr Res 1996; 20:105-115Crossref, Medline, Google Scholar
35. Pulver AE, Mulle J, Nestadt G, Swartz KL, Blouin J-L, Dombroski B, Liang K-Y, Housman DE, Kazazian HH, Antonarakis SE, Lasseter VK, Wolyniec PS, Thornquist MH, McGrath JA: Genetic heterogeneity in schizophrenia: stratification of genome scan data using co-segregating related phenotypes. Mol Psychiatry 2000; 5:650-653Crossref, Medline, Google Scholar
36. Mimmack ML, Ryan M, Baba H, Navarro-Ruiz J, Iritani S, Faull RL, McKenna PJ, Jones PB, Arai H, Starkey M, Emson PC, Bahn S: Gene expression analysis in schizophrenia: reproducible up-regulation of several members of the apolipoprotein L family located in a high-susceptibility locus for schizophrenia on chromosome 22. Proc Natl Acad Sci USA 2002; 99:4680-4685Crossref, Medline, Google Scholar
37. Lander E, Kruglyak L: Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 1995; 11:241-247Crossref, Medline, Google Scholar



