Confirmation That a Specific Haplotype of the Dopamine Transporter Gene Is Associated With Combined-Type ADHD
Abstract
Objective: The primary purpose of this study was to confirm the association of a specific haplotype of the dopamine transporter gene and attention deficit hyperactivity disorder (ADHD), which could be one source of the heterogeneity seen across published studies. Method: The authors previously reported the association of ADHD with a subgroup of chromosomes containing specific alleles of two variable-number tandem repeat polymorphisms within the 3′ untranslated region and intron 8 of the dopamine transporter gene. They now report on this association in a sample of ADHD combined-type probands. Results: The original observations were confirmed, with an overall odds ratio of 1.4 across samples. Conclusions: These data challenge results of meta-analyses suggesting that dopamine transporter variation does not have an effect on the risk for ADHD, and they indicate that further investigation of functional variation in the gene is required.
Attention deficit hyperactivity disorder (ADHD) is one of the most prevalent and heritable behavioral disorders of childhood. The disorder is characterized by onset of age-inappropriate hyperactivity, impulsivity, and inattentiveness before the age of 7 years. A familial risk has been established, and twin studies demonstrate the important role of genetic factors, with heritability estimates in the range of 60%–90%. Efforts to identify genes associated with ADHD have been relatively fruitful by focusing on genes that regulate dopamine and related monoamine neurotransmitter systems (1) .
One of the most prominent findings in ADHD research has been the association between the 10-repeat allele of a 40-base-pair variable-number tandem repeat (VNTR) in the 3′-untranslated (3′UTR) region of the dopamine transporter gene (DAT1). Although the association has been reported in multiple studies since the initial report by Cook et al. in 1995 (2) , there have also been many nonreplications (see references 1 , 3 , and 4 for reviews). Meta-analyses of available data have found the net effect across studies to be very small (3) , and one recent study suggested no effect at all (5) . However, several authors have also noted that the DAT1 data show significant evidence of heterogeneity across Caucasian data sets (5 – 7) . The reason for this heterogeneity is not well understood.
One possible contribution to heterogeneity between data sets might occur if the 10-repeat allele was tagging a nearby functional variant in partial linkage disequilibrium (LD) with the 10-repeat allele, with different levels of LD occurring in different populations. Alternatively, the 10-repeat allele may confer risk for ADHD only in combination with additional DNA variants in the DAT1 gene, and the specific combination might occur at different frequencies in various populations. Data supporting the role of alternative functional sites within DAT1 came from the recent study (8) of two distinct ADHD samples from the United Kingdom and Taiwan in which the association with ADHD was specific to a particular haplotype. The risk haplotype identified in the study consisted of the 10-repeat allele in the 3′UTR in conjunction with the six-repeat allele (which in our previous report we referred to as the three-repeat allele) of a 30-base pair VNTR located within intron 8 (8) . Since we know now that the repeat units are three more than originally reported, we have altered the allele numbering system to reflect this change; for example, the five-repeat allele was previously reported as a two-repeat allele and the six-repeat allele as a three-repeat allele. Of the four possible common haplotype combinations of the two markers (9-5, 9-6, 10-5, 10-6), only the 10-6 haplotype was associated with increased risk for ADHD in both populations.
Since then we have further investigated this key finding in a large sample of combined-subtype ADHD probands collected for the International Multicenter ADHD Gene (IMAGE) project.
Method
Descriptions of the clinical sample and the genotype assays are reported in detail elsewhere (9) . In brief, the sample consists of European Caucasian subjects recruited from 11 specialist clinics in eight countries: Belgium, Germany, Holland, Ireland, Israel, Spain, Switzerland, and the United Kingdom. All ADHD probands and their siblings were 5 to 17 years of age at the time of entry into the study, and access to one or both biological parents was required. Entry criteria for probands were a clinical diagnosis of DSM-IV combined-subtype ADHD and having one or more full siblings available for ascertainment of clinical information and DNA collection. Exclusion criteria applying to both probands and siblings included autism, epilepsy, IQ <70, brain disorders, and any genetic or medical disorder associated with externalizing behaviors that might mimic ADHD. A standardized algorithm was applied to data from the Parent Account of Childhood Symptoms, a semistructured investigator-based interview developed to provide objective measures of child behavior (10) , to derive each of the 18 DSM-IV ADHD items. These are combined with items that scored 2 or 3 from the ADHD subscale of the Conners’s Teacher Rating Scale (11) to generate the total number of items from the DSM-IV symptom checklist. Situational pervasiveness was defined as some symptoms occurring in two or more different situations from the Parent Account of Childhood Symptoms interview or the presence of one or more symptoms scoring 2 or more from the ADHD subscale of the Conners Teacher Rating Scale.
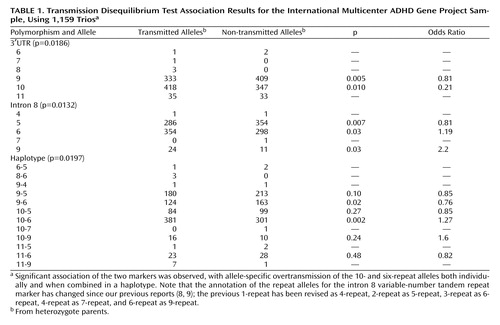
The final sample used in this study consisted of 998 families containing a total of 1,159 DSM-IV combined-type ADHD probands. DNA from both parents was available for 82.6% of probands (N=957) and from one parent for 13.8% (N=160). Most of the probands (86.6%, N=998) were male; 53.4% (N=620) had comorbid oppositional defiant disorder, and 24.3% (N=282) had conduct disorder; and 72.9% (N=845) were receiving medication for ADHD at the time of the research evaluations. Our analysis was conducted with the transmission disequilibrium test implemented in the software application UNPHASED (http://www.mrc-bsu.cam.ac.uk/personal/frank/software/unphased/). Genotype frequencies are available from the authors.
Results
We have already reported results from the first set of 776 probands from this sample (9) , in which we found that, similar to the initial U.K. and Taiwanese samples, only the 10-6 haplotype was overtransmitted from heterozygote parents to affected offspring (haplotype-specific p<0.06, odds ratio=1.19). We now find a similar pattern of findings in the second set of 383 ADHD probands from the IMAGE sample (haplotype-specific p<0.002, odds ratio=1.52). The evidence for the haplotype association in the IMAGE dataset is therefore very strong ( Table 1 ). Overall we find global evidence for association with the 3′UTR VNTR (p=0.019), the intron 8 VNTR (p=0.013), and the haplotype of the two markers (p=0.020). Examination of the allele-specific associations shows that for the individual markers, it is the 10-repeat and six-repeat alleles that are associated with risk for ADHD. More specifically, only the 10-6 combination is overtransmitted to ADHD offspring, whereas all other haplotype combinations are undertransmitted (haplotype-specific p=0.002, odds ratio=1.27), confirming the previous observation of a haplotype-specific association. The only possible exception is the observation in this sample that the rare nine-repeat allele from the intron 8 VNTR might also be overtransmitted to affected offspring.
Discussion
We are not aware of other available ADHD datasets that have genotyped the intron 8 VNTR together with the 3′UTR VNTR. The overall evidence to date combining the IMAGE data with the previously published data (8) for the 10-6 haplotype association shows 484 transmissions versus 341 nontransmissions (global p=4.26×10 –5 , haplotype-specific p=6×10 –7 , odds ratio=1.4), with none of the other three common haplotypes showing overtransmission to affected offspring.
Although in the combined data set we found associations of ADHD to both the 3′UTR VNTR (p=0.0005) and the intron 8 VNTR (p=0.0002), the important observation is that of the four possible common haplotype combinations, only the 10-6 haplotype is overtransmitted to affected offspring. This indicates that neither the 10-repeat allele nor the six-repeat allele alone can explain the association. To formally test this hypothesis, an alternative transmission disequilibrium test program, Whap (available at http://pngu.mgh.harvard.edu/~purcell/whap/), was used to investigate the effect of one marker conditional on the other. We compared the null hypothesis that the 10-6 haplotype is associated with ADHD and the alternative hypotheses of association with either the 10-repeat or the six-repeat allele alone. Dropping either VNTR from the analysis led to a significant drop in significance of the haplotype association (3′UTR p=0.002, intron 8 p=0.005), indicating that both markers contribute significantly to the haplotype association. Several other groups have identified haplotype-specific associations with the 10-repeat allele in combination with another allele from a second marker (12 – 14) , so the finding of haplotype-specific associations in this region is not unique to this study.
There are two main interpretations of these data. First, the two markers tag an as-yet unidentified causal variant that is in partial LD with the two markers. Second, there is a direct interaction between the two VNTR markers (or other causal variants in strong LD with the VNTR markers). LD is a measure of the correlation between marker genotypes on the same chromosome and can be very high for markers with similar allele frequencies that lie close together. In the IMAGE sample, we found that LD between the two markers was moderate to low (D′=0.42, R 2 =0.25) and was similar to that seen in the U.K. and Taiwanese samples (8) . The partial LD explains why the haplotype is more informative than the 3′UTR marker alone, since only a subset of chromosomes with the 10-repeat allele are associated with ADHD, whereas if LD were 0.8 or above, we would expect the intron 8 and 3′UTR markers to merely tag each other. Since the marker genotypes are correlated to a moderate degree, we cannot distinguish between the marker haplotype tagging a nearby functional variant and a two-locus interaction.
Taken together, these data challenge the results of meta-analyses that suggest no effect of DAT1 variation on risk for ADHD and indicate that further investigation of functional variation across the gene is required. In the most recent meta-analysis, Li et al. (6) concluded that a negative result is conclusive only if the entire gene has been tagged and there is sufficient power and uniformity in the studies. Other sources of heterogeneity, such as phenotypic differences between samples and interactions with environmental variables, may also be important but have yet to be systematically investigated.
1. Asherson P, IMAGE Consortium: Attention deficit hyperactivity disorder in the post-genomic era. Eur J Child Adoles Psych 2004; 13(suppl 1):I50–I70Google Scholar
2. Cook EH, Stein MA, Krasowski MD, Cox NJ, Olkon DM, Kieffer JE, Leventhal BL: Association of attention deficit disorder and the dopamine transporter gene. Am J Hum Genet 1995; 56:993–998Google Scholar
3. Faraone SV, Perlis RH, Doyle AE, Smoller JW, Goralnick JJ, Holmgren MA, Sklar P: Molecular genetics of attention-deficit/hyperactivity disorder. Biol Psychiatry 2005; 57:1313–1323Google Scholar
4. Thapar A, O’Donovan M, Owen MJ: The genetics of attention deficit hyperactivity disorder. Hum Mol Genet 2005; 14(spec no 2):275–282Google Scholar
5. Curran S, Mill J, Tahir E, Kent L, Richards S, Gould A, Huckett L, Sharp J, Batten C, Fernando S, Ozbay F, Yazgan Y, Simonoff E, Thompson M, Taylor E, Asherson P: Association study of a dopamine transporter polymorphism and attention deficit hyperactivity disorder in UK and Turkish samples. Mol Psychiatry 2001; 6:425–428Google Scholar
6. Li D, Sham PC, Owen MJ, He L: Meta-analysis shows significant association between dopamine system genes and attention deficit hyperactivity disorder. Hum Mol Genet 2006; 15:2276–2284Google Scholar
7. Maher BS, Marazita ML, Ferrell RE, Vanyukov MM: Dopamine system genes and attention deficit hyperactivity disorder: a meta-analysis. Psychiatr Genet 2002; 12:207–215Google Scholar
8. Brookes KJ, Mill J, Guindalini C, Curran S, Xu X, Knight J, Chen CK, Huang YS, Sethna V, Taylor E, Chen W, Breen G, Asherson P: A common haplotype of the dopamine transporter gene associated with attention-deficit/hyperactivity disorder and interacting with maternal use of alcohol during pregnancy. Arch Gen Psychiatry 2006; 63:74–81Google Scholar
9. Brookes K, Xu X, Chen W, Zhou K, Neale B, Lowe N, Aneley R, Franke B, Gill M, Ebstein R, Buitelaar J, Sham PC, Cambell D, Knight J, Andreou P, Altink M, Arnold R, Boer F, Buschgens C, Butler L, Christiansen H, Feldman L, Fleischman K, Fliers E, Howe-Forbes R, Goldfarb A, Heise A, Gabriëls I, Lubetzki I, Marco R, Medad S, Minderaa R, Mulas F, Müller U, Mulligan A, Rabin K, Rommelse N, Sethna V, Sorohan J, Uebel H, Psychogiou L, Weeks A, Barrett R, Craig I, Banaschewski T, Sonuga-Barke E, Eisenberg J, Kuntsi J, Manor I, McGuffin P, Miranda A, Oades RD, Plomin R, Roeyers H, Rothenberger A, Sergeant J, Steinhausen H-C, Taylor E, Thompson M, Faraone SV, Asherson P: The analysis of 51 genes in DSM-IV combined type attention deficit hyperactivity disorder: association signals in DRD4, DAT1, and 16 other genes. Mol Psychiatry 2006; 11:934–953Google Scholar
10. Taylor E, Schachar R, Thorley G, Wieselberg M: Conduct disorder and hyperactivity, I: separation of hyperactivity and antisocial conduct in British child psychiatric patients. Br J Psychiatry 1986; 149:760–767Google Scholar
11. Werry JS, Sprague RL, Cohen MN: Conners’ Teacher Rating Scale for use in drug studies with children: an empirical study. J Abnorm Child Psychol 1975; 3:217–229Google Scholar
12. Barr CL, Xu C, Kroft J, Feng Y, Wigg K, Zai G, Tannock R, Schachar R, Malone M, Roberts W, Nothen MM, Grunhage F, Vandenbergh DJ, Uhl G, Sunohara G, King N, Kennedy JL: Haplotype study of three polymorphisms at the dopamine transporter locus confirms linkage to attention/deficit-hyperactivity disorder. Biol Psychiary 2001; 49:333–339Google Scholar
13. Galili-Weisstub E, Levy S, Frisch A, Gross-Tsur V, Michaelovsky E, Kosov A, Meltzer A, Goltser T, Serretti A, Cusin C, Darvasi A, Inbar E, Weizman A, Segman RH: Dopamine transporter haplotype and attention deficit hyperactivity disorder. Mol Psychiatry 2005; 10:617–618Google Scholar
14. Hawi Z, Lowe N, Kirley A, Gruenhage F, Nothen M, Greenwood T, Kelsoe J, Fitzgerald M, Gill M: Linkage disequilibrium mapping at DAT1, DRD5, and DBH narrows the search for ADHD susceptibility alleles at these loci. Mol Psychiarty 2003; 8:299–308Google Scholar