Genomic Discordance Between Monozygotic Twins Discordant for Schizophrenia
Abstract
OBJECTIVE: Genomic DNA of monozygotic twins discordant for schizophrenia was analyzed to determine whether their genomes were truly identical. METHOD: The subjects were monozygotic male twins, one of whom had DSM-III-R schizophrenia, undifferentiated type. Genomic DNA was extracted from leukocytes and was applied to restriction landmark genome scanning analysis, which was developed for a high-speed survey of restriction sites throughout a genome and measurement of their copy number in each locus. RESULTS: After comparisons of patterns with approximately 2,000 spots, the authors detected at least two spots with autoradiographic intensities that obviously differed in the two twins. CONCLUSIONS: The discrepancies likely were generated either by differences in the methylation status at NotI sites between the twins or by submicroscopic changes occurring at NotI-flanking sites in one twin after (or simultaneous with) twinning. In either case, the difference may influence the transcription level of one or more genes.
Monozygotic twins discordant for schizophrenia provide clues to the etiological roles of environmental factors, because monozygotic twins are thought to be genetically identical (1, 2). However, it has been shown that certain somatic mutations occurring simultaneous with or after twinning may cause phenotypic differences between monozygotic twins (3, 4): monozygotic twins who are discordant for chromosomal abnormalities and for fragile X syndrome with different triplet repeat expansions in FMR1. Moreover, recent studies (3, 4) demonstrated that loss of genomic imprinting or skewed X inactivation in one twin may also cause a discordance. Thus, a method for identifying a genomic difference between monozygotic twins who are discordant for a disease can be a powerful tool for research on its etiology. Here we report differences of genomic DNA between monozygotic twins who are discordant for schizophrenia. The difference was detected by using a new technique, restriction landmark genome scanning (RLGS).
METHOD
The subjects were 44-year-old male twins who were entered in the Nagasaki Twin Registry. Twin A has suffered from DSM-III-R schizophrenia, undifferentiated type, since age 30 years, and has been treated by one of us (Y.O.) at Nagasaki University Hospital. He first developed a catatonic stupor and then 2 years later developed hallucinations of voices conversing. He is confined indoors without a job and never got married. Twin B is healthy and has never married, either. Although twin A did not continue his education after junior high school, twin B graduated from high school and has been entrusted with quality control in his employer's workshop for a long time. It was concluded that he did not have any psychiatric problems after psychiatric interviews by one of us (Y.O.). As normal comparison subjects, 28-year-old clinically healthy male twins were selected. After complete description of our study to the subjects, written informed consent was obtained. All the twins had 46,XY karyotypes without any visible chromosomal mosaicism. Monozygosity of the twins was proven by using seven blood types and DNA fingerprinting.
Genomic DNA was extracted from peripheral blood leukocytes of twins A and B and from those of the normal twin pair, and the DNA was analyzed with restriction landmark genome scanning, as described in detail elsewhere (5). In brief, genomic DNA (3.5 µg) was first treated with DNA polymerase I in the presence of deoxyguanosine [alpha-thio] triphosphate (dGTP[α]S), deoxycytidine [alpha-thio] triphosphate (dCTP[α]S), dideoxyadenosine triphosphate (ddATP), and dideoxythymidine triphosphate (ddTTP). This step, which is based on the incorporation of the dideoxynucleotide analogue into the nonspecifically damaged sites, such as nicks, gaps, or double strand breaks, is done to prevent background. The DNA was then digested with a landmark restriction enzyme, NotI, and the cleavage ends were end-labeled with a fill-in reaction by using Sequenase version 2.0 (U.S. Biochemical Corp., Cleveland, Ohio) in the presence of radiolabeled dCTP (6,000 Ci/mmol) and dGTP (3,000 Ci/mmol). The end-labeled DNA was digested with another enzyme, PvuII, and the DNA was electrophoresed in a 0.8% agarose gel (first dimension). The disc of the gel containing the DNA was treated with the third enzyme, PstI, followed by two-dimensional electrophoresis in 5% polyacrylamide gel. The gels were dried and exposed to Kodak XAR X-ray films at –80°C for several days.
RESULTS
Restriction landmark genome scanning revealed approximately 2,000 spots on each twin's autoradiogram. A detailed comparison of the patterns revealed at least two spots (spot 1 and spot 2) that showed obviously different autoradiographic intensities in twins A and B (figure 1). Both of the spots, estimated to be about 300–400 base pairs, were observed in twin A, while in twin B, spot 1 was lacking and spot 2 was only faintly visible (figure 1). The discrepancies were ascertained consistently by repeated trials of restriction landmark genome scanning. There was no visible difference in patterns between the normal comparison twins.
DISCUSSION
Restriction landmark genome scanning, developed for a high-speed survey of restriction sites throughout the genome, employs direct end-labeling of genomic DNA digested with a restriction enzyme and high-resolution, two-dimensional electrophoresis. As a spot's intensity accurately reflects the dose of a NotI/PstI restriction fragment, it is evident that twin A's genomic DNA contains two NotI/PstI fragments (spots 1 and 2), while the fragments are present in lower levels or are lacking in twin B. Since about three-fourths of monozygotic twins have in utero vascular connections (3, 4), it is reasonable that twin B has a faint spot 2. The reason why no additional spots corresponding to spot 1 or 2 were visible anywhere in twin B's autoradiogram is most likely the limited resolution power of restriction landmark genome scanning, by which only DNA fragments that are 70–2,000 base pairs long can be detected. Differences derived from variable immunoresponse system genes are generally undetectable, because the number of such individual cells to be subjected to restriction landmark genome scanning is negligible.
Two alternative mechanisms by which the discrepant fragments in the twins appeared are possible. First, as we used the methylation-sensitive enzyme NotI, the methylation status at one or more NotI sites may be different in the two twins. Although previous studies did not support an association between the etiology of schizophrenia and genomic imprinting (6), it is reasonable to assume the occurrence of epigenetic DNA modifications, such as methylation/demethylation, to explain the similar high rates of schizophrenia in the children of schizophrenic monozygotic twins and those of nonaffected co-twins (1). Usually, methylation/demethylation is reset in the next generation. Second, in either twin A or B, a submicroscopic change of DNA, e.g., deletion, insertion, or translocation, may have occurred at one or more NotI-flanking sites after (or simultaneous with) twinning. By either mechanism, since NotI sites frequently exist in CpG islands near the promoters of genes (7), the fragments have a high probability of reflecting such a mutation, leading to an alteration of the transcription level. In fact, from a NotI-linking clone library, several genes have successfully been isolated (8). Thus, it remains to be seen whether the genomic discordance observed in our monozygotic twins truly reflects different susceptibility to the disease. We are now trying to clone the discrepant DNA fragments with the gel-punching-out method (9).
This new approach makes it possible to detect directly and rapidly different DNA fragments in monozygotic twins. Such a difference results from a postzygotic event. However, it seems meaningless to compare autoradiogram spots from restriction landmark genome scanning of unrelated individuals because of the presence of numerous restriction-fragment-length polymorphisms in the genome. In other words, DNA fragments with different sizes at the same locus on the genome cannot generally be identified in unrelated individuals. Moreover, since schizophrenia is a polygenic disorder and shows genetic heterogeneity, it would not be surprising if other monozygotic twins discordant for the disease had identical patterns in restriction landmark genome scanning.
Received March 4, 1997; revision received Aug. 18, 1997; accepted Aug. 27, 1997. From the Department of Human Genetics and Department of Neuropsychiatry, Nagasaki University School of Medicine, and the Laboratory of Animal Breeding and Genetics, School of Agriculture, Kyushu Tokai University, Kumamoto, Japan. Address reprint requests to Dr. Tsujita, Department of Human Genetics, Nagasaki University School of Medicine, Sakamoto 1-12-4, Nagasaki 852, Japan; [email protected] (e-mail). Dr. Okazaki was supported by Grants-in-Aid for Scientific Research (B) from the Ministry of Education, Science, Sports, and Culture of Japan (grant 08457250) and by a Research Grant for Nervous and Mental Disorders from the Ministry of Health and Welfare of Japan. The authors thank Dr. Y. Hayashizaki at Riken Tsukuba Life Science Center for technical advice.
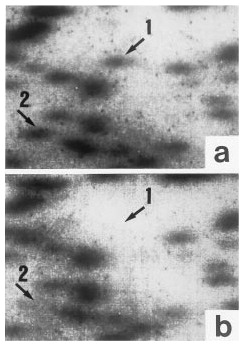
FIGURE 1. Differences in Patterns Shown by Restriction Landmark Genome Scanning Between Monozygotic Twins Discordant for Schizophreniaa
aSpot 1 is visible in the scan for the schizophrenic twin, twin A (a), but not in that for twin B (b). The intensity of spot 2 for twin B is weaker than that for twin A.
1. Gottesman II, Shields J: Schizophrenia: The Epigenetic Puzzle. New York, Cambridge University Press, 1982Google Scholar
2. Kendler KS: Overview: a current perspective on twin studies of schizophrenia. Am J Psychiatry 1983; 140:1413–1425Google Scholar
3. Hall JG: Twinning: mechanisms and genetic implications. Curr Opin Genet Dev 1996; 6:343–347Crossref, Medline, Google Scholar
4. Machin GA: Some causes of genotypic and phenotypic discordance in monozygotic twin pairs. Am J Med Genet 1996; 61:216–228Crossref, Medline, Google Scholar
5. Okazaki Y, Okuizumi H, Sasaki N, Ohsumi T, Kuromitsu J, Hirota N, Muramatsu M, Hayashizaki Y: A genetic linkage map of the mouse using an expanded production system of restriction landmark genomic scanning (RLGS Ver.1.8). Biochem Biophys Res Commun 1994; 205:1922–1929Google Scholar
6. Asherson P, Walsh C, Williams J, Sargeant M, Taylor C, Clements A, Gill M, Owen M, McGuffin P: Imprinting and anticipation: are they relevant to genetic studies of schizophrenia? Br J Psychiatry 1994; 164:619–624Google Scholar
7. Hattori M, Toyoda A, Ichikawa H, Ito T, Ohgushi H, Oishi N, Kano T, Kuhara S, Ohki M, Sasaki Y: Sequence-tagged NotI sites of human chromosome 21: sequence analysis and mapping. Ge~nomics 1993; 17:39–44Crossref, Google Scholar
8. Shimizu K, Ichikawa H, Tojo A, Kaneko Y, Maseki N, Hayashi Y, Ohira M, Asano S, Ohki M: An ets-related gene, ERG, is rearranged in human myeloid leukemia with t(16;21) chromosome translocation. Proc Natl Acad Sci USA 1993; 90:10280–10284Google Scholar
9. Ohsumi T, Okazaki Y, Hirotsune S, Shibata H, Muramatsu M, Suzuki H, Taga C, Watanabe S, Hayashizaki Y: A spot cloning method for restriction landmark genomic scanning. Electrophoresis 1995; 16:203–209Crossref, Medline, Google Scholar



