Predicting Depression
Innate predisposition to depression is both moderately heritable and influenced by early life experience. In this issue of the Journal, Halldorsdottir and colleagues (1) use a composite genotype “score” together with measures of child abuse to ask whether genetic (G) and environmental (E) risk factors interact. The authors explore the power of predictive testing at this early point in our understanding of the genetics of depression. To do this, they studied three cohorts of patients: two that were epidemiologic and a smaller but more deeply characterized clinical sample. Together, genes and environment acted additively, and rather than either superadditively or subadditively, in accordance with true G×E interaction.
These results are not the last word on genes, environment, and G×E models in depression because the assessment of genetic risk, based on a depression polygenic risk score (PRS) from a genome-wide association study (GWAS), captured less than 5% of the heritability of depression (2). Secondly, the depression phenotype, while clinically relevant, is nonspecific. Depression also occurs in specific contexts; for example, during changes in season, after traumatic events, during the premenstrual period, and during the postpartum period, as well as in chronic pain and addiction. As is inherent to the definitions of these conditions as diseases or their diagnostic exclusions in DSM-5, each of these varieties of depression could be marked by greater specificity of genes and G×E interaction. Clinically, contexts of depression can also point to more specific interventions and to, for example, the treatment of addiction or posttraumatic stress disorder (PTSD), the correction of an endocrine abnormality, or the relief of pain. Any of these more specific interventions can augment cognitive, behavioral, pharmacologic, and psychophysical therapies that are partly effective across the spectrum of depressive illnesses.
Potentially, genetic loci that are not well interrogated in GWASs or that in any case are not represented in a depression PRS derived from a GWAS could display true G×E interactions. An example of a G×E interaction relevant to depression, and involving a functional locus, is FKBP5, which puts the brakes on cellular response to cortisol. As discovered by Elisabeth Binder, the senior author of the Halldorsdottir et al. article, FKBP5 alters vulnerability to PTSD and major depression in the context of prior stress exposure (3). SLC6A4, the gene encoding the serotonin transporter, has been strongly connected to anxiety, depression, response to antidepressant drugs, and modulation of emotional circuits, including in animal models. The ability of a molecularly well-validated functional polymorphism at SLC6A4 (4) to increase depression and anxiety was thought to be conditional on stress exposure. However, a recent meta-analysis revealed that this polymorphism and stress are individually significant but act additively (5). Replication studies and meta-analyses have at times replicated (5) and at other times have failed to replicate (6) the main effects of SLC6A4. The failure of depression GWASs to detect a gene such as SLC6A4 may say a great deal about the power of a GWAS to detect genes’ effects, but it also drives questions of the nature and actions of genes implicated by depression GWASs. It is worthwhile to ask how the loci so far detected by GWASs modulate risk, even if we do not understand their nature, and it is imperative to know if depression PRSs, or a future, more informative depression PRS, might be delivered direct to consumer or disclosed physician to patient (Figure 1).
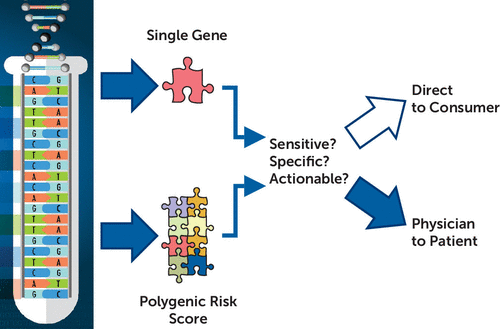
FIGURE 1. Criteria and process for introduction of genetic tests based on single genes or polygenic risk scorea
a Partly adapted from the National Human Genome Research Institute (https://www.genome.gov).
When is a genetic test or combined genetic-environmental predictor ready? At least three elements are needed: predictive power (measured as sensitivity and specificity), actionability, and a sound plan for delivering the information (Figure 1). In their clinical sample, Halldorsdottir et al. found that a depression PRS alone explained about 5% of the variance in case-control status in major depression, and with the additive effect of child abuse, somewhat less than one-fifth of the variance was explained. Moreover, it is also clear that PRS scores derived from psychiatric diseases predict risk of several diseases, each of which may require different treatment or prevention, if an intervention is available. The environmental risk factor studied by Halldorsdottir et al., child abuse, is also nonspecific, increasing odds of a variety of psychiatric diseases, including affective illness, anxiety disorders, addictions, and PTSD (7).
Actionable genetic predictors are available for many rare, Mendelian diseases. For these diseases, molecular diagnosis points to effective early interventions and can also spare patients from ineffective and dangerous treatments. As of July 2018, the U.S. Department of Health and Human Services recommends neonatal testing for 35 “core conditions,” including metabolic disorders such as phenylketonuria, endocrine disorders such as primary congenital hyperplasia, blood disorders such as sickle cell anemia, and multisystem disorders such as cystic fibrosis (https://www.hrsa.gov/advisory-committees/heritable-disorders/rusp/index.html). Such genetic tests are generally ordered by physicians for communication physician to patient, as is advisable given the complexities of interpretation, even though the results are likely to become quickly accessible by patients in their electronic health record. However, practically any genetic test is also available direct to consumer. Notably, the precision and actionability of a test do not necessarily lead to application. For example, testing for number of copies of the X and Y chromosomes would be informative for many diseases that are influenced by sex, including depression and breast cancer, but sex testing is not performed as such prenatally except in specific contexts, such as concern for X-linked diseases. Postnatally, sex can usually be determined by physical examination, obviating the need for a genetic test.
The challenge for psychiatry is prediction and diagnosis of common, non-Mendelian diseases. With the notable exception of large genetic insertions or deletions causing a substantial fraction of autism and psychosis, psychiatric diseases are not predicted with adequate sensitivity or specificity, although testing for depression, addictions, and other psychiatric diseases has been offered by several companies (both direct to consumer and physician to patient) for more than a decade. However, whatever may be the uptake by the medical community, there is little doubt that large-scale direct-to-consumer testing for psychiatric diseases is coming, and it will be based on PRSs because of the lack of informativeness of individual genes. This year, 23andMe, acknowledged in the Halldorsdottir et al. study, began reporting a diabetes PRS to its customers. 23andMe uses the diabetes PRS to inform individuals at higher risk (20% of the population) of their odds of this disease. Furthermore, people “at risk” are linked to a health coaching app, which is commercially available from a 23andMe partner. In 2013, the Food and Drug Administration (FDA) prohibited other polygenic predictions by 23andMe. However, in this instance, the FDA may agree that a diabetes PRS is innocuous, promoting general wellness activities that everyone should follow anyway. Perhaps the greater problem is that some people with favorable diabetes PRSs may feel reassured that they can skip regular exercise or eat poorly.
Will PRS-informed testing for depression take hold? Like type II diabetes, a large percentage of the population is vulnerable to depression, and depression is at least as heritable, and a PRS is readily calculable. For both diseases, there are early signs that can alternatively be used to instigate behavioral change: body mass index or serum triglyceride level for diabetes; decline in mood or interests in life’s rewards for depression. However, interventions to prevent the onset of depression are, if anything, more nonspecific. All children should be protected from abuse. Nearly everyone benefits by adopting aspects of healthy living that can prevent depression, whether they are genetically at risk. Scientifically derived, but otherwise not so different than a cleverly crafted horoscope, depression PRSs might be not so much a determinative guidepost for prevention as an innocuous reminder to live better. Even if a depression PRS is not highly predictive, as is presently true, careful consideration should be given to how such information would be communicated to the emotionally vulnerable.
1 : Polygenic risk: predicting depression outcomes in clinical and epidemiological cohorts of youths. Am J Psychiatry 2019; 176:615–625Link, Google Scholar
2 : Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet 2018; 50:668–681Crossref, Medline, Google Scholar
3 : Understanding the molecular mechanisms underpinning gene by environment interactions in psychiatric disorders: the FKBP5 model. Biol Psychiatry 2018; 83:821–830Crossref, Medline, Google Scholar
4 : Serotonin transporter promoter gain-of-function genotypes are linked to obsessive-compulsive disorder. Am J Hum Genet 2006; 78:815–826Crossref, Medline, Google Scholar
5 : Collaborative meta-analysis finds no evidence of a strong interaction between stress and 5-HTTLPR genotype contributing to the development of depression. Mol Psychiatry 2018; 23:133–142Crossref, Medline, Google Scholar
6 : No support for historical candidate gene or candidate gene-by-interaction hypotheses for major depression across multiple large samples. Am J Psychiatry 2019; 176:376–387Link, Google Scholar
7 : Prevalence, characteristics, and impact of childhood sexual abuse in a southwestern American Indian tribe. Child Abuse Negl 1997; 21:769–787Crossref, Medline, Google Scholar



