Reproducible Genetic Risk Loci for Anxiety: Results From ∼200,000 Participants in the Million Veteran Program
Abstract
Objective:
Anxiety disorders are common and often disabling. The goal of this study was to examine the genetic architecture of anxiety disorders and anxiety symptoms, which are also frequently comorbid with other mental disorders, such as major depressive disorder.
Methods:
Using one of the world’s largest biobanks including genetic, environmental, and medical information, the Million Veteran Program, the authors performed a genome-wide association study (GWAS) of a continuous trait for anxiety (based on score on the Generalized Anxiety Disorder 2-item scale [GAD-2], N=199,611) as the primary analysis and self-report of physician diagnosis of anxiety disorder (N=224,330) as a secondary analysis.
Results:
The authors identified five genome-wide significant signals for European Americans and one for African Americans on GAD-2 score. The strongest were on chromosome 3 (rs4603973) near SATB1, a global regulator of gene expression, and on chromosome 6 (rs6557168) near ESR1, which encodes an estrogen receptor. The locus identified on chromosome 7 (rs56226325, MAF=0.17) near MAD1L1 was previously identified in GWASs of bipolar disorder and schizophrenia. The authors replicated these findings in the summary statistics of two major published GWASs for anxiety, and also found evidence of significant genetic correlation between the GAD-2 score results and previous GWASs for anxiety (rg=0.75), depression (rg=0.81), and neuroticism (rg=0.75).
Conclusions:
This is the largest GWAS of anxiety traits to date. The authors identified novel genome-wide significant associations near genes involved with global regulation of gene expression (SATB1) and the estrogen receptor alpha (ESR1). Additionally, the authors identified a locus (MAD1L1) that may have implications for genetic vulnerability across several psychiatric disorders. This work provides new insights into genetic risk mechanisms underpinning anxiety and related psychiatric disorders.
Anxiety disorders are common, affecting 1 in 10 Americans each year, and are a leading cause of disability worldwide (1). An analysis of health expenditures in the United States found that anxiety and depressive disorders together accounted for about $90 billion in personal health spending in the United States in 2013 (2). Given their prevalence, associated impairment, and economic costs, anxiety disorders are a major public health concern (3).
Anxiety is “a future-oriented mood state associated with preparation for possible, upcoming negative events” (4) and is usually a normal and adaptive behavioral response to everyday life. In anxiety disorders, anxiety is excessive or out of proportion to the actual or anticipated event and is accompanied by clinically significant distress or disability (5). Numerous risk factors for anxiety disorders have been studied, including experiential and genetic factors (6). For example, neurotic personality traits are predictive of the onset of anxiety disorders (7). Twin studies demonstrate a heritable component to anxiety disorders (6), but there have been few published genome-wide association studies (GWASs) to date investigating anxiety or anxiety-related traits. The Anxiety Neuro Genetics Study (ANGST) (8) meta-analysis was the first large GWAS to identify significant genetic associations, finding one genome-wide significant locus each for a categorical case-control design for any anxiety disorder diagnosis and a quantitative factor score for anxiety in a cohort of over 18,000 subjects. Another recent large GWAS, from the Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH), identified a significant genetic association with anxiety and stress-related disorders in a cohort of 31,880 individuals in the national Danish registers (9). Also of note is a study based on the UK Biobank cohort, the second largest GWAS of anxiety to date (10), which examined anxiety using case-control and clinical cutoffs based on a score ≥10 on the Generalized Anxiety Disorder 7-item scale (GAD-7). While progress is being made, understanding of the genetics of anxiety disorders has lagged behind other related disorders, such as major depression (11).
Only a third of individuals with anxiety disorders receive treatment (12). For those who do enter treatment, psychological approaches such as cognitive-behavioral therapy (CBT) have been shown to be effective (13), as have certain pharmacotherapies (14). A recent systematic review of CBT treatment response rates for anxiety disorders showed average rates of 49.5% at end of treatment and 53.6% at follow-up (15). A better understanding of genetic risk factors and determinants now informs other aspects of medicine, such as oncology and cardiology, through identification of causal mutations (16) and variants, and this approach will have important implications for psychiatry (17). These precision-medicine approaches are challenging in complex traits such as anxiety, which are associated with many (perhaps hundreds of thousands of) variants of individually small effect (18). The use of polygenic risk scores will require a suitably large sample size to provide sufficient confidence in these small individual effects that cumulatively account for so much of the heritability (19). Underlying polygenic risk factors from sufficiently large cohorts may inform an approach to identifying individuals with a predisposition to anxiety disorders and to improving outcomes.
The Million Veteran Program (MVP), one of the world’s largest biobanks including genetic, environmental, and medical information, is based on data from U.S. military veterans (20–22). Using this large genetic data set and the Generalized Anxiety Disorder 2-item scale (GAD-2) (23) as well as self-reported physician diagnosis of an anxiety disorder, we discovered novel genome-wide significant variants associated with anxiety in European Americans and African Americans. We examined replication and genetic overlap of these results with previous studies of anxiety and traits with which anxiety disorders are commonly comorbid—major depression, PTSD, and neuroticism. We also examined expression quantitative trait loci (eQTLs) to identify possible gene expression implications of these genetic variants, with eQTL evidence for altered expression in the basal ganglia and cerebellum. These findings, in the largest cohort of individuals analyzed by GWAS for anxiety and anxiety disorders (199,611 subjects for the quantitative trait, 224,330 for binary self-report diagnosis) to date, indicate shared genetic risk with some other mental disorders but also point to loci that may be especially important for anxiety and anxiety-related traits.
Methods
Participants
The MVP cohort has been described previously (20). Results were analyzed in two separate tranches based on when genotyping results were available. Ancestry was assigned using 10 principal components and the 1000 Genomes Project phase 3 EUR and AFR data as reference within each MVP tranche.
Genotyping, Imputation, and Quality Control
Genotyping, imputation, and quality control within MVP has been previously described. Briefly, samples were genotyped using a 723,305-SNP Affymetrix Axiom Biobank array, customized for MVP (20). Imputation was performed with minimac3 using data from the 1000 Genomes Project. For postimputation quality control, SNPs with an imputation INFO score <0.3 or a minor allele frequency (MAF) <0.001 were removed from analysis. For the first tranche of data, 22,183 SNPs were selected through linkage disequilibrium (LD) pruning using PLINK 2.0 (24), and then Eigensoft (25) was used to conduct principal component analysis on 343,286 MVP samples and 2,504 1000 Genomes samples. The reference population groups (EUR, EAS, AFR, AMR, or SAS) in the 1000 Genomes samples were used to define European American (N=241,541) and African American (N=61,796) groups used in this analysis. Similar methods were used in the second tranche of data, which contained 108,416 new MVP samples and the same 2,504 1000 Genomes samples. In tranche 2, 80,694 participants were defined as European American and 20,584 as African American.
Phenotypic Assessment
We used the GAD-2 (23) for our primary analysis. The GAD-2 consists of two questions (see Table S1 in the online supplement) in a self-report survey, each scored on a scale of 0–3. Participants are asked to respond according to their symptoms during the past 2 weeks. Values for the two responses are summed, resulting in a range of scores between 0 and 6, which we treated as a continuous trait (Table 1). Mean GAD-2 scores in European American men (N=163,470) and women (N=11,693) were 1.08 (SD=1.64) and 1.64 (SD=1.87), respectively, and mean scores in African American men (N=21,153) and women (N=3,295) were 1.57 (SD=10.21) and 1.94 (SD=10.64), respectively. The mean ages of the European American and African American participants were 66.58 years (SD=11.62) and 60.6 years (SD=10.78), respectively.
| European Americans | African Americans | |||||||
|---|---|---|---|---|---|---|---|---|
| Age | Age | |||||||
| GAD-2 Score | N | Mean | SE | Female (%) | N | Mean | SE | Female (%) |
| 0 | 100,141 | 69.10 | 0.033 | 4.79 | 11,212 | 62.93 | 0.1 | 10.64 |
| 1 | 21,569 | 65.31 | 0.082 | 8.20 | 2,897 | 60.33 | 0.2 | 15.15 |
| 2 | 25,061 | 63.95 | 0.075 | 8.86 | 3,947 | 58.97 | 0.166 | 16.06 |
| 3 | 8,450 | 62.40 | 0.129 | 9.44 | 1,708 | 58.86 | 0.243 | 14.87 |
| 4 | 8,046 | 61.49 | 0.134 | 10.08 | 1,730 | 57.83 | 0.253 | 16.99 |
| 5 | 4,447 | 60.61 | 0.176 | 9.83 | 1,011 | 57.30 | 0.333 | 15.23 |
| 6 | 7,449 | 59.16 | 0.143 | 11.63 | 1,943 | 56.40 | 0.235 | 16.83 |
| Total | 175,163 | 66.58 | 0.028 | 6.68 | 24,448 | 60.59 | 0.069 | 13.48 |
TABLE 1. Subjects and phenotype distribution in a study of genetic risk loci for anxietya
Another anxiety phenotype—self-reported physician diagnosis of anxiety disorder—was analyzed based on data collected from the MVP baseline survey. Participants were asked, “Please tell us if you have been diagnosed with the following conditions: anxiety reaction/panic disorder.” Answers were recorded as yes/no binary responses, and missing responses were excluded from analysis. A total of 224,330 participants (34,189 case subjects who responded yes, 190,141 control subjects who responded no) had available phenotype and genotype information and had assignments of either European ancestry (28,525 cases, 163,731 controls) or African ancestry (5,664 cases, 26,410 controls) (see Table S2 in the online supplement).
Statistical Analysis
GWAS analysis was carried out by linear regression for each ancestry group and tranche using PLINK 2.0 on genotype dosage data, covarying for age, sex, and the first 10 principal components against the phenotype of GAD-2 score. Ancestry-specific and trans-ancestry meta-analysis were performed using inverse variance weighting in the METAL software package (European American, N=175,163; African American, N=24,448; combined trans-ancestry, N=199,611). Logistic regression was used for self-reported physician diagnosis of an anxiety disorder, and the results obtained were combined using the same meta-analytic approach. To identify independent GWAS signals, we clumped results using an r2 of 0.10 and window size of 1,000 kb. Post-GWAS analyses were conducted for what turned out to be the most genetically informative phenotype based on z-scored heritability: GAD-2 score.
Linkage Disequilibrium Score Regression and SNP-Based Heritability
We used linkage disequilibrium score regression through LD Hub (26) to estimate SNP-based heritability and to assess genetic correlation of GAD-2 anxiety with all traits available in LD Hub. The traits from the ANGST GWAS of anxiety case-control and factor scores (8) and iPSYCH anxiety and stress-related disorders (9)—neither of which were available in LD Hub—were calculated separately with LD score regression software (LDSC) using summary statistics downloaded from the Psychiatric Genomics Consortium (PGC) web site (https://www.med.unc.edu/pgc/results-and-downloads/) or from the authors, respectively.
Conditional Analysis for Major Depression
Considering the extensive comorbidity between major depression and anxiety disorders (6), we ran a conditional analysis with the multi-trait-based conditional and joint analysis method (mtCOJO) (27) using the GCTA software package. This method uses GWAS summary statistics from one trait to perform conditional analysis on GWAS summary statistics from another trait. We conditioned the MVP GAD-2 summary statistics as the primary analysis with the PGC major depressive disorder (11) summary statistics for depression. We quantified changes in variance explained by using LD score regression to calculate heritability in the depression-conditioned GAD-2 analysis and compared with the original GAD-2 analysis.
Gene-Based Tests
Summary statistics from the GWAS were loaded into the FUMA (Functional Mapping and Annotation) GWAS platform to test for gene-level associations using Multi-Marker Analysis of GenoMic Annotation (MAGMA) (28). Input SNPs were mapped to 18,469 protein coding genes. The genome-wide significance threshold for the gene-based test was defined in accordance with Bonferroni multiple testing correction (p=0.05/18,469=2.71×10−6).
Fine Mapping
Fine mapping was conducted using PAINTOR, version 3 (29). A brain functional annotation set (30) was used to prioritize causal SNPs. The z-scored GAD-2 GWAS summary statistics served as the base analysis data set, with the aforementioned brain data set serving as the functional annotation. We enumerated all possible combinations and searched for a single causal SNP within each locus.
Results
Primary Analysis
GWAS of GAD-2 scores was conducted separately in two tranches of each ancestry in the MVP sample, defined by the time when data became available, and meta-analyzed together within ancestral group. One genomic locus was genome-wide significant in the African American meta-analysis (Figure 1A), and five loci were genome-wide significant in the European American meta-analysis (Figure 1B). The genome-wide significant result from the African American analysis (rs575403075, MAF=0.06, p=2.82×10−8) was near the TRPV6 (Transient Receptor Potential Cation Channel Subfamily V Member 6) locus. The top signal in the European American meta-analysis consisted of 64 genome-wide significant SNPs in high LD at the SATB1-AS1 (Special AT-Rich Sequence Binding 1 Antisense RNA 1) locus on chromosome 3. The strongest finding (rs4603973, MAF=0.29, p=6.18×10−11) was intronic at SATB1-AS1. The second strongest independent signal was on chromosome 6 (rs6557168, MAF=0.37, p=1.33×10−9) intronic at ESR1 (Estrogen Receptor 1) with 10 other genome-wide significant SNPs in high LD. A third genome-wide significant association for European Americans was found on chromosome 1 (rs12023347, MAF=0.48, p=8.88×10−9) near the long noncoding RNA LINC01360 and LRRIQ3 (Leucine-Rich Repeats and IQ motif containing 3). The fourth genome-wide significant association found in European Americans was on chromosome 7 (rs56226325, MAF=0.17, p=2.01×10−8) in an intron of MAD1L1 (Mitotic Arrest Deficient 1 Like 1). The fifth association for European Americans was on chromosome 20 in and around the TCEA2 (Transcription Elongation Factor A2), RGS19 (Regulator of G Protein Signaling 19), and OPRL1 (Opioid Related Nociceptin Receptor 1) genes (rs6090040, MAF=0.48, p=3.28×10−8).
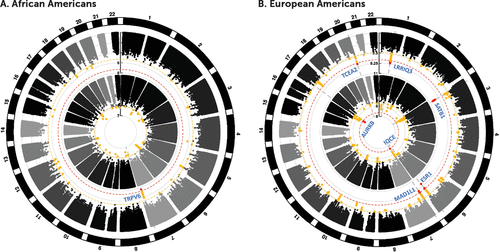
FIGURE 1. Circle Manhattan plot for anxiety phenotypes in African Americans and European Americansa
a The outer circle displays results of the Generalized Anxiety Disorder 2-item scale genome-wide association study (GWAS), and the inner circle contains results for the case-control (self-report of physician diagnosis of anxiety disorder) GWAS. Numbers outside the circle represent chromosomes. Red dots indicate genome-wide significant findings (p<5 x10−8) and yellow dots indicate suggestive findings (p<5×10−6). The scale on the y-axis represents −log10(p value). Vertical dashed gray lines are drawn through genome-wide significant findings to indicate overlap between analyses. The genes nearest to the lead SNP are labeled adjacent to the result. In most cases a genome-wide significant (red) locus from one phenotype overlaps with at least a suggestive (yellow) locus in the other.
We conducted additional analyses using case-control status for self-reported physician diagnosis of an anxiety disorder. For the European American subjects, there were two genome-wide significant signals for the GWAS of self-reported physician diagnosis of an anxiety disorder, in a gene-rich region nearest AURKB on chromosome 17 (rs35546597, MAF=0.42, p=1.88×10−8) and on chromosome 7 in an IQCE intron (rs10534613, MAF=0.41, p=4.92×10−8) close to the MAD1L1 locus identified for GAD-2. There were no genome-wide significant findings for this phenotype in African Americans.
Replication
For replication, we tested our top five SNPs from the analysis of GAD-2 scores in European Americans in three independent GWASs with anxiety-related phenotypes. We considered a replication to be significant if the p value was <0.05. We investigated our lead genome-wide significant SNPs in GWASs for the ANGST anxiety case-control (8), iPSYCH anxiety and stress-related disorders (9), and UK Biobank, 23andMe, and Genetics of Personality Consortium (GPC) neuroticism (31) phenotypes (Table 2). The first two phenotypes are very similar to our GAD-2 measure; the third is best considered a related phenotype (rg=0.7174, p=1.95×10−53).
| GAD-2 MVP European American Meta-Analysisb | iPSYCH Anxiety and Stress-Related Disorders (9) | UK Biobank, 23andMe, and GPC Neuroticism (31) | ANGST Anxiety (8) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RSID | CHR | Gene | p | Risk Allele | p | Risk Allele | p | Risk Allele | p | Risk Allele |
| rs4603973 | 3 | SATB1-AS1 | 6.18E–11 | G | 0.181 | G | na | na | 0.0299 | G |
| rs4390955 | 3 | SATB1-AS1 | 7.78E–11 | A | 0.851 | C | 5.20E–04 | A | 0.2935 | A |
| rs6557168 | 6 | ESR1 | 1.33E–09 | C | 0.0128 | C | 0.367 | C | 0.170 | C |
| rs12023347 | 1 | LINC01360 /LRRIQ3 | 8.88E–09 | T | 6.61E–04 | T | 6.85E–04 | T | 0.00296 | T |
| rs56226325 | 7 | MAD1L1 | 2.01E–08 | C | 6.41E–04 | C | 6.59E–08 | C | 0.354 | C |
| rs6090040 | 20 | TCEA2 | 3.28E–08 | C | 0.461 | A | 0.444 | A | 0.867 | C |
TABLE 2. Replication in independent cohorts in a study of genetic risk loci for anxietya
In the ANGST anxiety study (8), which was the smallest replication cohort, all five of our top independent SNPs had the same direction of effect, with two being nominally significant (p<0.05). In the iPSYCH study of anxiety and stress-related disorders (9), four of five independent SNPs had the same direction of effect, with three being nominally significant (p<0.05). We also replicated the lead SNP from iPSYCH near PDE4B in our own study (iPSYCH lead SNP: rs7528604, p=5.39×10−8; present study GAD-2: same SNP, p=0.015). Only one of our findings, near OPRL1, failed to replicate in at least one independent study.
Our lead SNP on chromosome 3 near the SATB1 locus, rs4603973, was not available for lookup in the neuroticism GWAS (31), which we used as a proxy replication of a related trait, so we used the strongest LD-proxy SNP available (rs4390955 R2=0.91, p=7.78E-11). In this study, four of our five independent SNPs we looked up had the same direction of effect, three were nominally significant (p<0.05), and one near MAD1L1 was nearly genome-wide significant (UK Biobank neuroticism: rs56226325, p=6.59×10−8; present study GAD-2: same SNP, p=2.01×10−8).
Lastly, a preprint reported results for anxiety from the UK Biobank using case-control and the GAD-7, scored as a dichotomous trait (10). We found significant replication for two of their four findings, with suggestive evidence for a third (see Table S9 in the online supplement).
Genome-Wide Gene-Based Association Study for GAD-2
In the genome-wide gene-based association study, the top gene identified was OPRL1 (p=1.15×10−9), which was also significant in the SNP-wise analysis, as noted above. Thirty-one genes were identified as genome-wide significant following Bonferroni correction for multiple comparisons. A more permissive Benjamini-Hochberg correction performed by step-up procedure, with genes ranked by p value and corrected for 18,469 individual tests with a still relatively restrictive 0.05 false discovery rate, identified 189 genes (see Table S3 in the online supplement) in total for investigation of biological relevance through the Ingenuity pathway enrichment tool (32), Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, Calif.; www.ingenuity.com) (see Table S4 in the online supplement). Among the top enriched diseases or functional annotations were carcinoma (p=1.76×10−7) and fear conditioning (p=3.62×10−4).
Expression Quantitative Trait Loci (eQTLs)
To identify causal implications for genetic variants, eQTLs were assessed for the top genome-wide significant GAD-2 signals using GTEx version 7 brain tissue expression data. Top genome-wide significant signals on chromosomes 7 and 20 had significant eQTLs (false discovery rate ≤0.05) for four different genes: FTSJ2, RGS19, C20orf201, and OPRL1 (see Table S7 in the online supplement). The top signals are centered in the basal ganglia and cerebellum.
SNP-Based Heritability
SNP-based heritability using LDSC for the GAD-2 quantitative trait was estimated to be 5.58% (SE=0.004, z-score=13.95). SNP-based inflation was mild considering the sample size and polygenic trait studied (λ=1.19); the intercept (1.026) and attenuation ratio (0.1177) estimated by LDSC showed negligible evidence for inflation due to population stratification. SNP-based heritability for the self-reported physician diagnosis of an anxiety disorder binary trait was 8.79% (SE=0.0085, z-score=10.34) on the liability scale assuming prevalence of 20%. This value is similar to that reported for anxiety by Otowa et al. (h2=0.095, SE=0.037, z-score=2.57) (8), depression by the PGC (h2=0.087, SE=0.004, z-score=21.75) (11) and Howard et al. (h2=0.089, SE=0.003, z-score=29.67) (33), and neuroticism by Nagel et al. (h2=0.100, SE=0.003, z-score=33.33) (31), but somewhat lower than that reported by Meier et al. for anxiety and stress-related disorders (h2=0.28, SE=0.027, z-score=10.37) (9).
Linkage Disequilibrium Score Regression Analysis
The traits most significantly genetically correlated with GAD-2 score were depressive symptoms (rg=0.81, p=1.95×10−53) and neuroticism (rg=0.72, p=6.53×10−53). We also investigated genetic correlation within the MVP cohort for GAD-2 score and self-reported physician diagnosis of an anxiety disorder. These were high (rg=0.87, p=2.39×10−119), and higher than the phenotypic correlation between these traits (r=0.64, p<2.2×10−16) (Figure 2; see also Table S5 in the online supplement).
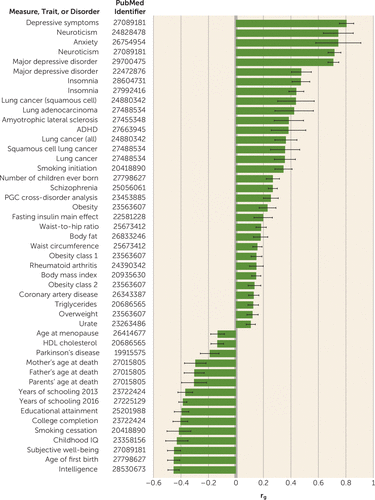
FIGURE 2. Genetic correlation between Million Veteran Program GAD-2 score in European Americans and other traits and disorders from LD score regression in LD Huba
a All plotted traits survive 0.05 false discovery rate. Full results are presented in Table S5 in the online supplement. GAD-2=Generalized Anxiety Disorder 2-item scale; LD=linkage disequilibrium; PGC=Psychiatric Genomics Consortium.
Polygenic Risk Score (PRS) Analysis
Summary statistics from the MVP GAD-2 analysis were used as the base data for calculating polygenic risk scores (PRSs) (using PRSice, version 1.25 [34]). Genetic overlap between anxiety and PTSD or major depressive disorder was tested using the “summary statistics to summary statistics” procedure, using the gtx R package incorporated into PRSice, in the PGC major depressive disorder (11) and PTSD (35) GWASs, respectively, and overlap with case-control anxiety disorder was tested in the largest previously available studies (8, 9). Significant overlap was identified: the MVP GAD-2 PRS can explain up to 0.24% of the variance in major depressive disorder in the PGC GWAS (p=2.05×10−94), 0.23% of the variance in PTSD in the PGC GWAS (p=4.23×10−12), and 0.48% of the variance in both the ANGST (p=3.66×10−20) and iPSYCH anxiety studies (p=6.68×10−36) (Table 3).
| Trait (Reference Number) | GWAS p Threshold | SNPs Tested (N) | r2 | p |
|---|---|---|---|---|
| PGC depression (1) | 0.145 | 89,152 | 0.002445 | 2.05E–94 |
| PGC PTSD (35) | 0.075 | 58,854 | 0.002330 | 4.23E–12 |
| ANGST anxiety (8) | 0.415 | 83,033 | 0.004884 | 3.66E–20 |
| iPSYCH anxiety (9) | 0.130 | 79,974 | 0.004853 | 6.68E–36 |
TABLE 3. Polygenic risk scores generated from the Million Veteran Program genome-wide association study (GWAS) of GAD-2 anxiety trait and predicting into other relevant GWAS summary statisticsa
Multi-Trait-Based Conditional and Joint Analysis
Multi-trait-based conditional and joint analysis was used to condition the GAD-2 MVP summary statistics for anxiety on the PGC summary statistics for major depressive disorder (11). There were no new signals and the significance levels of the lead findings were reduced, but the results on chromosomes 3 (SATB1) and 6 (ESR1) remained genome-wide significant. Degree of lost variance explained in the anxiety GWAS when conditioned on major depressive disorder was tested using LD score regression. Genetic correlation analysis was performed between the original European American GAD-2 GWAS summary statistics and the major depressive disorder conditioned summary statistics, which served as an internal control to show that the trait measured was still the same (rg=1.0). Heritability dropped significantly (p=0.021) from 0.0558 (SE=0.0041) in the original GWAS to 0.0429 (SE=0.0038) in the conditioned GWAS.
Fine Mapping
Fine mapping in PAINTOR, version 3, was used to predict causal SNPs using functional brain annotations (see Figure S6 in the online supplement). In one case (chromosome 6, rs6557168) the causal SNP identified was the same as the GWAS lead SNP. On chromosome 20, there were several genes in the region of our lead SNP, and several genes had associated eQTLs. The fine mapping analysis prioritized a likely causal SNP (rs8126001) within the 5′ UTR of OPRL1.
Discussion
We present the largest GWAS to date for anxiety traits, employing a quantitative phenotype, the GAD-2 score, in nearly 200,000 MVP subjects, as well as self-reported physician diagnosis of anxiety/panic case-control phenotypes in >220,000 MVP subjects. We identified novel genetic variants in and around several genes, some of which have previously known functional relationships with anxiety. These genes play roles in the hypothalamic-pituitary-adrenal (HPA) axis, neuronal development, and global regulation of gene expression.
There is high comorbidity between anxiety, PTSD, and depression. We used a PRS derived from the MVP GAD-2 analysis to identify genetic overlap with the independent PGC PTSD and major depressive disorder GWASs (Table 3; see also Figure S7 in the online supplement). We found significant genetic overlap between these traits, providing biological evidence that this known clinical comorbidity is due at least in part to shared genetic etiology. Additionally, we performed multi-trait-based conditional and joint analysis, using a prior GWAS of depression to condition the results of the present GAD-2 GWAS. In this analysis, we show not only that the peak signals for anxiety are reduced in magnitude (see Figure S3 in the online supplement) but also that the overall heritability for anxiety symptoms is diminished, from 5.58% to 4.29%, when conditioned on genetic liability to depression. Via linkage disequilibrium score regression, we identified substantial genetic correlations between anxiety and numerous other traits (Figure 2). Particularly noteworthy were positive correlations with depression and neuroticism as well as a negative correlation with subjective well-being (Figure 2). These findings replicate similar correlations found using a case-control approach (9).
The genome-wide significant result in African Americans is an insertion variant that is rare outside of African ancestry and occurs in a genomic region proposed to be under recent selection in Europeans (36). The lead SNP is at TRPV6, which encodes a Ca2+-selective membrane cation channel associated with epithelial calcium transport and homeostasis in kidney and intestine. The lead SNP rs575403075 has an MAF range of between 0% and 1% in non-African populations and would fall below MAF quality control thresholds used for common variants in most non-African populations. In individuals of African ancestry, this variant is much more common, with an MAF in our study of 5.8%. This highlights the importance of studying genetic risks in diverse populations—otherwise these signals may be missed entirely.
The top genome-wide significant findings for European Americans in the GAD-2 analysis were in and around SATB1 and the antisense gene SATB1-AS1. SATB1 is a global regulator that influences expression of multiple genes involved in neuronal development (37). One gene modulated in expression is Corticotropin Releasing Hormone (CRH), encoding the protein product of the same name that plays an essential role in the HPA axis, which has frequently been shown to modulate stress and fear/anxiety response (38). The CRHR1 (Corticotropin Releasing Hormone Receptor 1) gene was genome-wide significant in the gene-based association analysis (p=3.60×10−7). CRHR1 has been a proposed target for treatment for anxiety and stress-related disorders, with evidence for anxiolytic-like effects of CRHR1 antagonists in animal models although not yet in humans. Based on our findings, we speculate that individuals with differing genetic risk that does or does not involve this pathway may differ in their responses to CRHR1-targeted and other glucocorticoid-targeted therapeutic agents; this may be a reasonable pathway to address via personalized medicine, and it presents a testable hypothesis.
The estrogen receptor ESR1 (also known as estrogen receptor alpha) has been a focus in animal models of anxiety-like behaviors, and these have provided mechanistic validity for the role of ESR1. Studies of estradiol administration to ovariectomized rats and ESR1 null mice have shown consistent evidence that ESR1 is involved in anxiety-like behavior (39). Our finding of an association between ESR1 and anxiety may have implications for our understanding of sex differences in anxiety disorders and trauma and stressor-related disorders such as PTSD, which are more common in females (40). Although this female predominance is partially explained by sex-specific exposure to certain kinds of traumatic events (e.g., domestic violence, sexual assault), there may also be differential biological context provided in part by the role of the estrogen receptor. Our study in a predominantly male sample identifies ESR1 as genome-wide significant. Estrogen is important in both sexes, and a recent review has highlighted the important role for estrogens in men (41). Studies with larger numbers of women will be needed to more fully investigate sex differences in genetic risk for anxiety-related traits.
Previous genetic epidemiology studies have shown that common genetic factors can underlie anxiety and depressive traits (42). The lead SNP from the GAD-2 GWAS near the LINC01360 and LRRIQ3 (rs2180945) loci is nominally significant and has the same direction of effect in the 2018 PGC major depressive disorder analysis (p=1.434×10−6) (11). This variant may be linked to a common risk factor for both disorders.
One genome-wide significant signal for GAD-2 was in a gene-rich region on chromosome 20 near TCEA2, C20orf201, RGS19, and OPRL1, with fine-mapping analysis prioritizing a causal region in the latter gene. OPRLI (which encodes the amygdala nociceptin/orphanin FQ receptor) is involved in learning and memory and anxiety and fear-related behaviors (43, 44) and has been hypothesized to play a role in anxiety and stressor-related disorders such as PTSD (44). Interestingly, fear conditioning was also significantly enriched in the pathway analysis. Taken together, these observations suggest that OPRL1 and related systems should be further explored as targets for anxiety and stressor-related therapeutics. eQTL data suggest that variants in this region regulate expression of RGS19 and OPRL1 in the cerebellum and in the basal ganglia (see Table S7 in the online supplement). The basal ganglia have long been implicated in obsessive-compulsive disorder and anxiety disorders. A recent review discussed cerebellum-linked neurocircuitry to anxiety and fear behaviors in rodents and in humans (45). The cerebellum is thought to play an important role in anticipation/prediction processes. Given that anxiety has been defined as “a future-oriented mood state associated with preparation for possible, upcoming negative events” (4), these results may provide further evidence for a role for the cerebellum in fear and anxiety.
MAD1L1 (GAD-2 lead SNP rs56226325, MAF=0.17, p=2.01×10−8; self-reported physician diagnosis of an anxiety disorder lead SNP rs10534613, MAF=0.41, p=4.92×10−8) is replicated in the iPSYCH anxiety GWAS data (9) (Table 2; p=6.85×10−4) and has been associated previously with bipolar disorder (46). One of the lead SNPs in the iPSYCH study is also nominally associated with anxiety in the present study (rs11764590, p=3.36×10−7); this SNP is in LD with our lead SNP, rs56226325 (r2=0.69). A recent large GWAS of bipolar disorder identified genome-wide significant SNPs in the MAD1L1 locus, although their lead signal is not significant in our study of anxiety (rs4236274, p=0.27) (47). This locus has also been identified among 108 genome-wide significant loci by the PGC schizophrenia study (rs58120505, p value=6.43×10−14) (48), and our lead SNP is nominally significant in that study (rs56226325, p=1.12×10−3). This SNP is also nominally significant (6.71×10−4) in the 2018 PGC depression GWAS (11). Taken together, these observations suggest that this locus may be a common risk factor for several psychiatric disorders.
MAD1L1*rs56226325 is also an eQTL for expression of FTSJ2 (see Table S7 in the online supplement), and this variant is associated with decreased expression in the brain. MAD1L1 is a mitochondrial RNA methyltransferase that is important for the proper assembly of the mitochondrial ribosome and cellular respiration (49). The protein product of FTSJ2 is Mitochondrial rRNA Methyltransferase 2 (MRM2), which was implicated in a case study of a 7-year-old Italian boy with a damaging mutation that reduced the catalytic activity of MRM2, leading to an encephalopathy, lactic acidosis, and stroke-like (MELAS) syndrome (50). Larger-effect mutations at this locus can have devastating effects on the brain; smaller-effect variations may be less deleterious but still cumulatively influence development, which may predispose to neurological and psychiatric disorders.
The Brainstorm Consortium has investigated shared heritability between psychiatric and neurological disorders (51). Consistent with their findings, we find very strong genetic correlation between anxiety (GAD-2) and psychiatric traits such as depression (rg=0.81, p=2.48×10−53) and neuroticism (rg=0.72, p=7.09×10−53) but relatively weaker genetic overlap with neurological disorders. Significant positive genetic association is detected for amyotrophic lateral sclerosis (rg=0.39, p=3.00×10−4), and negative genetic association with Parkinson’s disease (rg=−0.19, p=4.70×10−3), but no genetic overlap is seen for Alzheimer’s disease (rg=0.00, p=1.00). Further work will be needed to better discern the implications of these findings for understanding shared and disparate disease mechanisms among these neuropsychiatric conditions (11, 35, 51).
Limitations of this work include the fact that phenotypes were based on self-reported survey data. The GAD-2 asks questions that temporally reference the “past 2 weeks.” Although the GAD-2 has demonstrated high sensitivity and specificity for anxiety disorders (52), it falls short of the desired trait (lifetime) anxiety measure. That our work reproduces (and is reproduced by) other independent groups who did use lifetime anxiety measures (8, 9) further supports the utility of the GAD-2 in capturing a genetically meaningful anxiety trait. Similarly, the question about diagnosis for anxiety or panic that yielded our binary self-reported physician diagnosis of anxiety or panic disorder phenotype relies on self-report. A further limitation is that MVP has predominantly male participants (92.5%). While women are included in this analysis, clinically important interactions between sex, phenotype, and genotype could not be addressed. This cohort is growing, and future recruitment will provide additional power to revisit sex-stratified analyses of this sample. Males presumably have a higher genetic liability threshold for anxiety, as evidenced by lower rates of anxiety disorders. Accordingly, affected males could reflect higher genetic risk than females (because they must pass a higher threshold to be affected), which would result in greater power to detect risk loci in a mostly male compared with a mostly female sample.
In summary, we have identified novel variants for anxiety by performing GWASs in the large MVP cohort. We replicated results in our GWASs for top findings from recent anxiety and other relevant anxiety-related GWASs. We also identified significant genetic overlap with major depressive disorder, PTSD, and neuroticism using polygenic risk scores and LD score regression. This work provides additional genetic evidence for the overlap between disorders that are frequently comorbid with anxiety and presents new molecular targets for investigation with a longer view toward the development of new treatments.
1 : Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018; 392:1789–1858Crossref, Medline, Google Scholar
2 : Mental disorders top the list of the most costly conditions in the United States: $201 billion. Health Aff (Millwood) 2016; 35:1130–1135Crossref, Medline, Google Scholar
3 : Treating Anxiety in 2017: optimizing care to improve outcomes. JAMA 2017; 318:235–236Crossref, Medline, Google Scholar
4 : What is an anxiety disorder? Depress Anxiety 2009; 26:1066–1085Crossref, Medline, Google Scholar
5 : Diagnostic and Statistical Manual of Mental Disorders, 5th ed (DSM-5). Washington, DC, American Psychiatric Association, 2013Crossref, Google Scholar
6 : Anxiety disorders. Nat Rev Dis Primers 2017; 3:17024Crossref, Medline, Google Scholar
7 : Testing a hierarchical model of neuroticism and its cognitive facets: latent structure and prospective prediction of first onsets of anxiety and unipolar mood disorders during 3 years in late adolescence. Clin Psychol Sci 2016; 4:805–824Crossref, Google Scholar
8 : Meta-analysis of genome-wide association studies of anxiety disorders. Mol Psychiatry 2016; 21:1391–1399Crossref, Medline, Google Scholar
9 : Genetic variants associated with anxiety and stress-related disorders: a genome-wide association study and mouse-model study. JAMA Psychiatry (Epub ahead of print, May 22, 2019)Google Scholar
10 : The common genetic architecture of anxiety disorders. bioRxiv, October 16, 2017 (
11 : Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet 2018; 50:668–681Crossref, Medline, Google Scholar
12 : The global burden of anxiety disorders in 2010. Psychol Med 2014; 44:2363–2374Crossref, Medline, Google Scholar
13 : Cognitive-behavioral and pharmacological treatments of social phobia: a controlled study. Arch Gen Psychiatry 1991; 48:938–945Crossref, Medline, Google Scholar
14 : Anxiety disorders, in American Psychiatric Association Publishing Textbook of Psychiatry, 7th ed. Edited by Roberts LW. Washington, DC, American Psychiatric Association Publishing, 2018, pp 341–370Google Scholar
15 : Response rates for CBT for anxiety disorders: need for standardized criteria. Clin Psychol Rev 2015; 42:72–82Crossref, Medline, Google Scholar
16 : Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat Genet 2017; 49:594–599Crossref, Medline, Google Scholar
17 : Precision psychiatry: will genomic medicine lead the way? JAMA Psychiatry 2018; 75:663–664Crossref, Medline, Google Scholar
18 : An expanded view of complex traits: from polygenic to omnigenic. Cell 2017; 169:1177–1186Crossref, Medline, Google Scholar
19 : What are polygenic scores and why are they important? JAMA 2019; 321:1820–1821Crossref, Medline, Google Scholar
20 : Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 2016; 70:214–223Crossref, Medline, Google Scholar
21 : Genome-wide association study of post-traumatic stress disorder reexperiencing symptoms in >165,000 US veterans. Nat Neurosci 2019; 22:1394–1401Crossref, Medline, Google Scholar
22 : Genome-wide association study of maximum habitual alcohol intake in >140,000 U.S. European and African American veterans yields novel risk loci. Biol Psychiatry 2019; 86:365–376Crossref, Medline, Google Scholar
23 : The Patient Health Questionnaire Somatic, Anxiety, and Depressive Symptom Scales: a systematic review. Gen Hosp Psychiatry 2010; 32:345–359Crossref, Medline, Google Scholar
24 : PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81:559–575Crossref, Medline, Google Scholar
25 : Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38:904–909Crossref, Medline, Google Scholar
26 : LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 2017; 33:272–279Crossref, Medline, Google Scholar
27 : Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun 2018; 9:224Crossref, Medline, Google Scholar
28 : Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017; 8:1826Crossref, Medline, Google Scholar
29 : Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet 2014; 10:
30 : Systematic localization of common disease-associated variation in regulatory DNA. Science 2012; 337:1190–1195Crossref, Medline, Google Scholar
31 : Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat Genet 2018; 50:920–927Crossref, Medline, Google Scholar
32 : Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014; 30:523–530Crossref, Medline, Google Scholar
33 : Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci 2019; 22:343–352Crossref, Medline, Google Scholar
34 : PRSice: Polygenic Risk Score software. Bioinformatics 2015; 31:1466–1468Crossref, Medline, Google Scholar
35 : Largest GWAS of PTSD (N=20 070) yields genetic overlap with schizophrenia and sex differences in heritability. Mol Psychiatry 2018; 23:666–673Crossref, Medline, Google Scholar
36 : Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol 2004; 2:
37 : Satb1 ablation alters temporal expression of immediate early genes and reduces dendritic spine density during postnatal brain development. Mol Cell Biol 2012; 32:333–347Crossref, Medline, Google Scholar
38 : Effects of antalarmin, a CRF type 1 receptor antagonist, on anxiety-like behavior and motor activation in the rat. Brain Res 2002; 952:188–199Crossref, Medline, Google Scholar
39 : Estrogen receptors modulation of anxiety-like behavior. Vitam Horm 2017; 103:27–52Crossref, Medline, Google Scholar
40 : Gender differences in rates of depression, PTSD, pain, obesity, and military sexual trauma among Connecticut war veterans of Iraq and Afghanistan. J Womens Health (Larchmt) 2010; 19:267–271Crossref, Medline, Google Scholar
41 : Estrogens in male physiology. Physiol Rev 2017; 97:995–1043Crossref, Medline, Google Scholar
42 : Genetic and environmental influences on the co-morbidity between depression, panic disorder, agoraphobia, and social phobia: a twin study. Depress Anxiety 2009; 26:1004–1011Crossref, Medline, Google Scholar
43 : Nociceptin and the nociceptin receptor in learning and memory. Prog Neuropsychopharmacol Biol Psychiatry 2015; 62:45–50Crossref, Medline, Google Scholar
44 : Activation of nociceptin/orphanin FQ receptors inhibits contextual fear memory reconsolidation. Neuropharmacology 2017; 125:39–49Crossref, Medline, Google Scholar
45 : The cerebellum in fear and anxiety-related disorders. Prog Neuropsychopharmacol Biol Psychiatry 2018; 85:23–32Crossref, Medline, Google Scholar
46 : Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am J Hum Genet 2011; 88:372–381Crossref, Medline, Google Scholar
47 : Genome-wide association study of 40,000 individuals identifies two novel loci associated with bipolar disorder. Hum Mol Genet 2016; 25:3383–3394Crossref, Medline, Google Scholar
48 : Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511:421–427Crossref, Medline, Google Scholar
49 : MRM2 and MRM3 are involved in biogenesis of the large subunit of the mitochondrial ribosome. Mol Biol Cell 2014; 25:2542–2555Crossref, Medline, Google Scholar
50 : Defective mitochondrial rRNA methyltransferase MRM2 causes MELAS-like clinical syndrome. Hum Mol Genet 2017; 26:4257–4266Crossref, Medline, Google Scholar
51 : Analysis of shared heritability in common disorders of the brain. Science 2018; 360:
52 : Anxiety disorders in primary care: prevalence, impairment, comorbidity, and detection. Ann Intern Med 2007; 146:317–325Crossref, Medline, Google Scholar



