Allele-Specific Methylation of SPDEF: A Novel Moderator of Psychosocial Stress and Substance Abuse
Abstract
Objective:
Psychosocial stress is a key risk factor for substance abuse among adolescents. Recently, epigenetic processes such as DNA methylation have emerged as potential mechanisms that could mediate this relationship. The authors conducted a genome-wide methylation analysis to investigate whether differentially methylated regions are associated with psychosocial stress in an adolescent population.
Methods:
A methylome-wide analysis of differentially methylated regions was used to examine a sample of 1,287 14-year-old adolescents (50.7% of them female) from the European IMAGEN study. The Illumina 450k array was used to assess DNA methylation, pyrosequencing was used for technical replication, and linear regression analyses were used to identify associations with psychosocial stress and substance use (alcohol and tobacco). Findings were replicated by pyrosequencing a test sample of 413 participants from the IMAGEN study.
Results:
Hypermethylation in the sterile alpha motif/pointed domain containing the ETS transcription factor (SPDEF) gene locus was associated with a greater number of stressful life events in an allele-dependent way. Among individuals with the minor G-allele, SPDEF methylation moderated the association between psychosocial stress and substance abuse. SPDEF methylation interacted with lifetime stress in gray matter volume in the right cuneus, which in turn was associated with the frequency of alcohol and tobacco use. SPDEF was involved in the regulation of trans-genes linked to substance use.
Conclusions:
Taken together, the study findings describe a novel epigenetic mechanism that helps explain how psychosocial stress exposure influences adolescent substance abuse.
Environmental stressors are linked to the onset of psychiatric symptoms (1), many of which emerge during adolescence (2). For example, repeated exposure to severe adversity—such as childhood maltreatment or peer victimization—is associated with increased risk for substance abuse in adolescents (3). Even more common psychosocial stressors (such as problems in relationships, problems at school, and parental divorce) can influence adolescents’ propensity to substance abuse later in life (4). Although the importance of environmental factors is well understood, the biological mechanisms that underlie their effects on substance abuse, and their genetic interactions, are poorly characterized.
Recently, variability in DNA methylation—an epigenetic process regulating gene expression (5)—has emerged as a potential mechanism by which environmental adversity could translate into vulnerability to developing substance use among adolescents (6). It has been established that DNA methylation is sensitive to environmental influences, such as psychosocial adversity (7), that DNA methylation can be affected by genetic influences, as demonstrated by allele-specific methylation (8), and that aberrant DNA methylation patterns are linked to a range of psychopathologies, including substance abuse (9). Together, these findings give rise to the hypothesis that the effect of environmental influences on psychiatric symptoms and disorders may be mediated by variations in DNA methylation. However, there is insufficient empirical evidence to confirm or reject this hypothesis.
Furthermore, while recent studies have reported altered methylation patterns of toll-like receptor 4 (TLR4) (10), monoamine oxidase (MAO) (11), and glucocorticoid receptor genes in relation to substance use disorders (12), including drinking and smoking, these studies mainly focused on candidate genes. A methylome-wide analysis, in contrast, can provide a hypothesis-free platform, thus potentially identifying genes that point toward novel biological pathways (13).
We therefore carried out a genome-wide methylation analysis of psychosocial stress in adolescents and investigated its relationship to substance abuse. By using data from the population-based IMAGEN study (14), we measured DNA methylation from whole blood, assessed the frequency of stressful life events in the past year, and quantified lifetime substance use. To analyze the association between stress and DNA methylation, we applied a genome-wide region-based approach (15) that increases sensitivity to detect CpG clusters with greater biological relevance by taking into account the high correlation of methylation profiles in neighboring CpG sites (16). Lastly, we investigated the effect of single-nucleotide polymorphisms (SNPs) on methylation, examined the relationship between methylation and cis-gene and trans-gene expression, and analyzed the relationship between methylation and brain structure.
Methods
Participants
A total of 1,700 participants from the IMAGEN cohort were randomly assigned to an exploration sample (N=1,287) or a test sample (N=413) with similar demographic characteristics for replication (female, 53.3%; mean age, 14.44 years [SD=0.45]). IMAGEN is a European multicenter imaging-genetics study of adolescents recruited from England, France, Ireland, and Germany (14). All data from the IMAGEN study are available online (https://imagen2.cea.fr).
Questionnaire and Interview Data
Negative stressful life events.
An adapted version of the Life Events Questionnaire (17), a 39-item tool, was used to assess life events of individuals in the IMAGEN cohort during adolescence (for further details, see the online supplement). Given that the neurobiological effects of stress depend on the age at onset, we focused on the frequency of negative stressful life events that occurred in the past year only as a measure of interest.
Substance use.
The European School Survey Project on Alcohol and Other Drugs questionnaire (18) was used to assess the frequency of lifetime smoking, lifetime alcohol use, and lifetime binge drinking (for further details, see the online supplement).
Methylation, Genetic, and Gene Expression Data
Methylation.
DNA was extracted from whole blood samples and bisulphite treated using the EZ 96 DNA methylation kit (Zymo Research, Irvine, Calif.). DNA methylation was quantified with the Illumina Infinium HumanMethylation450 BeadChip (19) run on an Illumina HiScan System (Illumina, San Diego) using the manufacturer’s standard protocol. Pyrosequencing was carried out for technical validation and independent replication with a PSQ96 genetic sequencer using PyroMark Gold Q96 reagents (Qiagen, Valencia, Calif.) in accordance with the manufacturer’s recommendations (for further details, see the online supplement).
Genetic.
Genotype data were collected for 582,982 markers using DNA extracted from whole blood, as previously described, as well as the Illumina HumanHap610 Genotyping BeadChip. Genotype data were coded as the number of major alleles (for details on quality control, see the online supplement).
Gene expression.
Total RNA was extracted from whole blood cells collected from participants at age 14 using the PAXgene Blood RNA Kit (Qiagen, Valencia, Calif.) (for further details, see the online supplement). Gene expression profiling was performed using Illumina HumanHT-12, version 4, Expression BeadChips (Illumina, San Diego).
MRI Data Acquisition and Preprocessing
Structural data were acquired at eight IMAGEN assessment sites with 3-T MRI scanners (Siemens, Munich; Philips, Amsterdam; and General Electric, Boston). Full details of MRI acquisition protocols and quality control checks, including standardization across MRI scanners, have been published elsewhere (14). Details of the preprocessing are provided in the online supplement.
Data Analysis
All analyses controlled for the effects of recruitment site, gender, and cell count (for further details, see the online supplement). Analyses involving life events controlled for emotional valence to ensure that effects were linked to frequency rather than subjective perception. Analyses involving methylation data were controlled for acquisition waves (for further details, see the online supplement). All analyses were performed with SPSS, version 20.0 (IBM, Armonk, N.Y. [20]) or R, version 3.1.3 (https://cran.r-project.org), unless indicated otherwise. For all analyses, a significance threshold set at <0.05 was applied. Pearson’s correlation coefficient (r) is reported, except as otherwise indicated.
Identification of differentially methylated regions.
Data collected using the Illumina Infinium HumanMethylation450 BeadChip were analyzed with the minfi package in the Bioconductor software suite (21). Methylation data underwent quality control (for further details, see the online supplement) and were preprocessed using the stratified quantile normalization implemented in minfi (21). The bumphunter function in minfi R (15) was used to identify clusters of neighboring CpGs differentially methylated with psychosocial stress (see the online supplement). Post hoc quality control was conducted to exclude SNPs located under the probe that may affect the stability of the probe’s hybridization and its extension efficiency (22).
Gene-environment interaction analysis.
A general linear model was used to examine between-subject effects of genotype, frequency of negative stressful life events, and their interaction with methylation. Effect size is reported as Cohen’s d.
Stress, methylation, and substance use.
Separate multiple regression models were used to establish the relationship between psychosocial stress, methylation, and substance use. A one-tailed test was used for the analyses between methylation and substance use. A total of 10,000 permutations were applied to correct for multiple testing.
Stress-methylation interaction analysis.
General linear models were used to examine the effect of the frequency of negative stressful life events, methylation, and their interaction with substance use and gray matter volumes independently.
Gray matter volume and substance use.
A group comparison between participants with a high frequency of combined drinking and smoking and participants who did not consume alcohol or nicotine was conducted using two-sample t tests with the significance threshold set at 0.05 (two-tailed).
Blood-brain methylation association.
The Blood Brain DNA Methylation Comparison Tool (http://epigenetics.iop.kcl.ac.uk/bloodbrain) was used to examine the consistency of methylation across blood and brain (23) (for further details, see the online supplement).
Methylation-expression quantitative trait loci analyses.
The methylation-expression quantitative trait loci (eQTL) model was performed using the Matrix eQTL R package (24), and the database for annotation, visualization, and integrated discovery (DAVID) (25) was used to determine enrichment of biological processes and disease classes (for further details, see the online supplement). Only individuals with both methylation and gene expression data were included in the analyses (N=277).
Intra- and interchromosomal interactions between the SPDEF and trans-genes.
To determine the physical associations between the SPDEF promoter region and the differentially expressed trans-genes, the Hi-C browser for SKNMC cells (http://promoter.bx.psu.edu/hi-c/index.html) was used to examine the intra- and interchromosomal interactions (26).
Results
Association of Differentially Methylated Regions With Frequency of Stressful Life Events
Genome-wide DNA methylation was assessed in 1,287 adolescents (50.7% of them female; mean age, 14.45 years [SD=0.63]) using the Illumina 450k array (Table 1). Genome-wide analysis was conducted to identify differentially methylated regions associated with the frequency of negative stressful life events (for further details, see the online supplement). We found a genome-wide significant differentially methylated region (p=4.16×10−6; family-wise error-corrected p=0.018) located in the 5′ region of the sterile alpha motif/pointed domain epithelial specific transcription factor (SPDEF) gene (Table 2). Within this SPDEF region, we identified two CpG sites (cg16527629 and cg01395541) that were both significantly associated with the frequency of stress (r=0.094, df=1201, p=0.001 and r=0.082, df=1201, p=0.004, respectively). These CpG sites contained SNPs rs2233632 (0 bp) and rs2233631 (2 bp), respectively (Figure 1). To validate the 450k data, we carried out pyrosequencing analysis for both CpG sites in 94 individuals and found high correlations of methylation levels (cg01395541: r=0.85, p=2.21×10−37; cg16527629: r=0.82, p=6.77×10−24) (for further details, see the online supplement) consistent with previous comparisons (27). An investigation of the SPDEF sequence revealed that the CpG site cg01395543 harbored the binding motif of the transcription factor YY1, GCAT/CCAT: TTGGTGGTGGGTACCTCTGTCCTGC[A/G]TGGCATCCCTGCCATCACCCTTTGG.
| Variable | N | Mean | SD | Minimum | Maximum |
|---|---|---|---|---|---|
| Behavioral data | |||||
| Frequency of negative stressful life events in the past year | 1,213 | 2.80 | 1.78 | 0 | 10 |
| Emotional valence | 1,213 | –16.65 | 4.62 | –27 | 4 |
| European School Survey Project on Alcohol and Other Drugs questionnaire | |||||
| Lifetime smoking | 1,275 | 0.87 | 1.70 | 0 | 6 |
| Lifetime alcohol use | 1,272 | 2.01 | 1.73 | 0 | 6 |
| Lifetime binge drinking | 1,272 | 0.63 | 1.24 | 0 | 5 |
| Genetic data | |||||
| Methylation | |||||
| Mean SPDEF | 1,287 | 0.45 | 0.26 | 0.10 | 0.83 |
| SPDEF cg16527629 | 1,287 | 0.34 | 0.24 | 0.06 | 0.84 |
| SPDEF cg01395541 | 1,287 | 0.57 | 0.28 | 0.13 | 0.92 |
| Single-nucleotide polymorphism | |||||
| Genotype | |||||
| rs2233632 | 1,112 | 1.37 | 0.66 | ||
| GG | 114 | ||||
| GA | 468 | ||||
| AA | 530 | ||||
| rs2233631 | 1,112 | 1.38 | 0.66 | ||
| GG | 114 | ||||
| GA | 467 | ||||
| AA | 531 | ||||
| Gene expression | |||||
| Illumina HumanHT–12 Version 4 Expression BeadChips | 660 | ||||
| Neuroimaging data | |||||
| T1 gray matter | 512 |
TABLE 1. Descriptive data for the study participants in a methylome-wide analysis of psychosocial stress and SPDEFa
| Allele-Specific Frequencies and Methylation Levels | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene (Gene Symbol) | Chr | p | Corrected p | Location | CpG Sites Located Within Differentially Methylated Region | SNPs | GG | GA | AA |
| Sterile alpha motif/pointed domain containing ETS transcription factor (SPDEF) | 6 | 4.16×10–6 | 0.018 | Promoter | Cg01395541 | rs2233631 | 8.9%, N=114, M=0.82, SD=0.02 | 36.3%, N=467, M=0.83, SD=0.04 | 41.3%, N=531, M=0.27, SD=0.08 |
| Cg16527629 | rs2233632 | 8.9%, N=114, M=0.77, SD=0.07 | 36.4%, N=468, M=0.48, SD=0.04 | 41.2%, N=530, M=0.10, SD=0.03 | |||||
| Hook microtubule tethering protein 2 (HOOK2) | 19 | 4.86×10–6 | 0.021 | Covers exons | Cg04657146, Cg06417478, Cg11738485, Cg23899408 | ||||
TABLE 2. Results from bump hunting in a methylome-wide analysis of psychosocial stress and SPDEF

FIGURE 1. Single-nucleotide polymorphisms rs2233632 and rs2233631 in the differentially methylated region within the SPDEF genea
a Configured using the Santa Cruz Genomics Institute Genome Browser, build GRCh37/hg19 (https://genome.ucsc.edu).
We identified an additional differentially methylated region in the hook microtubule-tethering protein 2 (HOOK2) gene but excluded it from further analyses because it contained SNPs located at the probe binding site, and this could have resulted in false DNA methylation signals (for further details, see the online supplement).
Genetic control of methylation levels.
To determine the effect of SNP genotype on methylation at each CpG site, we examined the allele-specific methylation levels. We found that methylation levels for GG and GA were higher than the levels for the frequent homozygotes (Table 2). In addition, we found that the frequency of alleles at rs2233632 and rs2233631 was significantly associated with methylation levels at the respective CpG sites cg16527629 and cg01395541 (r=−0.982, p<2.2×10−16 and r=−0.876, p<2.2×10−16). Thus, the main association previously observed between stress and methylation was at least in part dependent on genotype.
Interaction between genotype, stress, and methylation.
Because methylation levels at CpG sites cg01395541 and cg16527629 are genotype dependent, we tested whether methylation was exclusively under genetic control or whether there was a gene-environment interaction with methylation at each CpG site. We found a significant interaction between the frequency of stressful life events and rs2233631 genotypes associated with methylation levels at cg01395541 (T=2.831, Cohen’s d=0.174, p=0.005, two-tailed), indicating that the level of methylation at cg01395541 was dependent on both genotype and stress exposure. An interaction was not observed for cg16527629 (p=0.338), indicating that the methylation levels observed were entirely the result of a SNP effect. We therefore excluded cg16527629 from further analysis.
When the SNP rs2233631 carries the G allele, the sequence (5′-TGCGTGGCAT-3′) contains a CpG site that can potentially be methylated. However, when rs2233631 carries the A allele, the CpG site is abrogated in the sequence (5′-TGCATGGCAT-3′), resulting in an absence of methylation. Hence, we repeated the analyses in the remaining SNP genotypes (GA and GG) (Figure 2A). We found that the effect size of the interaction between the frequency of stressful life events and rs2233631 genotypes (GA and GG) with methylation levels at cg01395541 increased by 18% (T=2.403, Cohen’s d=0.205, p=0.017, two-tailed) upon removal of AA homozygotes.
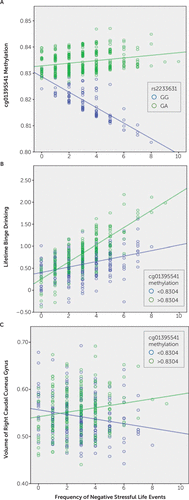
FIGURE 2. Results of interaction analysesa
a Panel A shows the interaction of the frequency of negative stressful life events and single-nucleotide polymorphism genotype with cg01395541 methylation. Panel B shows the interaction of the frequency of negative stressful life events and cg01395541 methylation with lifetime frequency of binge drinking. Panel C shows the interaction of the frequency of negative stressful life events and cg01395541 with gray matter volume in the right caudal cuneus gyrus.
Because DNA methylation is known to be a dynamic epigenetic mark, we also investigated the effect of lifetime stress on cg01395541 methylation. We did not find a significant interaction of lifetime stress and genotype with methylation (T=1.420, p=0.156), suggesting that cg01395541 methylation was due to the accumulation of stress within the past year and not compounded by lifetime stress exposure.
Independent replication of interaction between stress and genotype with methylation.
To replicate our interaction findings, we used bisulfite pyrosequencing to assess methylation levels for SPDEF cg01395541 in a test sample of 413 individuals from the IMAGEN study. We replicated the interaction between stress and rs2233631 SNP genotypes (GA and GG) with methylation levels at the CpG site cg01395541 (T=2.239, p=0.013). A meta-analysis of the exploratory and replication samples showed an interaction between stress and SNP genotype with methylation levels at cg01395541 (Z=3.195, p=0.0014, two-tailed).
Integrating SPDEF Methylation, Stress, and Substance Use
Associations between stress and substance use.
To investigate the relationship between psychosocial adversity and substance abuse in the IMAGEN sample, we examined the association between stress and substance use in all genotypes. We found that the frequency of negative stressful life events was positively associated with lifetime smoking (r=0.21, r2=0.0441, p=2.07×10−13), lifetime alcohol consumption (r=0.16, r2=0.0256, p=1.64×10−8), and lifetime binge drinking (r=0.14, r2=0.0196, p=6.81×10−7).
Stratifying by genotype, we found stronger associations between the frequency of negative stressful life events and lifetime alcohol consumption (r2=0.0303, p=5.3×10−5) and lifetime binge drinking (r2=0.0234, p=3.92×10−4) in carriers of the G allele compared with AA homozygotes, suggesting that G-allele carriers are more susceptible to developing alcohol abuse as a result of psychosocial stress exposure. No such difference was observed for lifetime smoking (r2=0.0437, p=1.04×10−6).
Associations between methylation and substance use.
Next, we sought to determine the relationship between methylation and substance use problems by assessing the association between methylation levels at site cg01395541 and drinking and smoking behaviors. Methylation levels at cg01395541 were positively associated with lifetime binge drinking (r=0.099, p=0.008; permutation-based p=0.0228), and an association with lifetime smoking fell just short of statistical significance (r=0.087, p=0.020; permutation-based p=0.0507); the association with lifetime alcohol consumption was not significant (p=0.268).
Interaction of stress and methylation with substance use.
Because we found separate main effects of stress and methylation on substance use, we hypothesized that cg01395541 methylation influenced the link between psychosocial stress and drinking and smoking behaviors. We observed a significant interaction between the frequency of negative stressful life events and cg01395541 methylation levels with lifetime binge drinking (T=2.274, df=541, p=0.023, two-tailed) (Figure 2B). When comparing the variance explained by the two models (the model with main effects only and the model including the interactions), we found that the model that included the interactions explained significantly more variance (partial r2=0.0256, p=0.00837), suggesting that methylation at cg01395541 moderates the association between stress and lifetime binge drinking, which is also demonstrated by a causal analysis (for further details, see the online supplement). No interaction was observed for lifetime smoking (p=0.118, two-tailed).
Interaction of stress and methylation with gray matter volume.
We examined the interaction of stress and methylation with brain structure and found an interaction of the frequency of negative stressful life events and cg01395541 methylation with gray matter volume in the right caudal cuneus gyrus (T=3.718, p=0.000225; p=0.0482, Bonferroni-corrected) (Figure 2C). This interaction was significant for both boys (T=2.268, p=0.024) and girls (T=2.840, p=0.005). Additionally, we found that this region was smaller in participants who engaged in a high frequency of smoking or drinking compared with those who did not consume alcohol or nicotine (t=−1.989, p=0.048).
Blood-brain methylation association.
To examine the consistency of SPDEF methylation across blood and brain, we used the online Blood Brain DNA Methylation Comparison Tool. Our blood-based methylation at site cg01395541 was highly correlated with methylation in all brain regions for which data were available, in particular the prefrontal cortex (r=0.991), entorhinal cortex (r=0.981), superior temporal gyrus (r=0.989), and cerebellum (r=0.989) (Figure 3). This suggests that methylation at cg01395541 was highly consistent across blood and brain tissues. However, we were not able to test this relationship experimentally, because there are insufficient numbers of adolescent postmortem brains available.
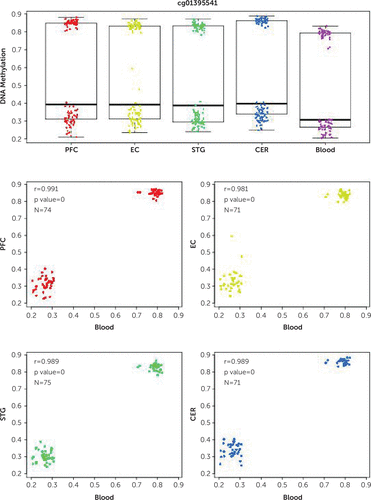
FIGURE 3. Correlation of methylation at the CpG site cg01395541 between blood and various brain regionsa
a Configured using the Blood Brain DNA Methylation Comparison Tool. CER=cerebellum, EC=entorhinal cortex, PFC=prefrontal cortex, STG=superior temporal gyrus.
SPDEF Methylation and Gene Expression
To explore the relationship between methylation at the CpG site cg01395541 and gene expression, we performed a methylation-eQTL analysis. Although there was no association between methylation at cg01395541 and SPDEF cis-gene expression, we found an association of methylation with expression of multiple trans-genes (for further details, see Table S2 in the online supplement). A total of 246 probes mapping 159 genes associated with methylation levels at cg01395541 for genotypes GG and GA were found (p<0.01) (for further details, see Table S2 in the online supplement).
Using DAVID, we found an overrepresentation of gene networks associated with opioid-related disorders (p=0.0023), alcoholism (p=0.012), and tobacco use disorder (p=0.037) (for further details, see Table S3 in the online supplement). The genes for the mu opioid (OPRM1) and dopamine (DRD2) receptors, two genes previously associated with substance use disorders, were present in all of these enriched pathways.
To assess the physical interaction between the SPDEF promoter region and the identified trans-genes, we conducted intra- and interchromosomal interactions between our SPDEF differentially methylated region and the genes listed in Table S2 in the online supplement using the Hi-C browser. We found that SPDEF was physically linked to 12 genes (for further details, see Figure S3 in the online supplement), including DRD2, present in gene networks associated with alcohol dependence and immune response, suggesting that SPDEF could be involved in the regulation of expression of these trans-genes.
Discussion
We conducted a genome-wide methylation study to investigate the effect of psychosocial stress exposure on DNA methylation in the population-based IMAGEN cohort of 14-year-old adolescents. We identified differential methylation in a novel gene, SPDEF, associated with psychosocial stress exposure. By dividing the IMAGEN cohort into an exploratory sample and a test sample (28), we showed and replicated an association of SPDEF methylation at the CpG site cg01395541 with lifetime binge drinking and lifetime smoking. We did not find an association between SPDEF methylation and lifetime alcohol consumption, likely because of the young age of the study participants, resulting in lifetime consumption being a limited measure: more than 60% reported consuming alcohol on less than 10 occasions in their lifetime. There was an interaction of SPDEF methylation and lifetime stress with gray matter volume in the right caudal cuneus. Cuneus volume was negatively correlated with the frequency of drinking and smoking. Furthermore, we found that SPDEF methylation moderated the relationship between psychosocial stress exposure and substance use. Our methylation-eQTL analysis suggests that SPDEF is involved in biological pathways linking psychosocial stress and substance abuse.
SPDEF is an ETS (E26 transformation-specific) transcription factor that has been previously linked to various cancers (29) and was reported to be an androgen-independent transactivator of prostate-specific androgen promoter (30). This study is the first, to our knowledge, to implicate SPDEF in stress response and substance abuse. The identified differentially methylated region was located in the promoter region of the gene, and further analysis on the sequence revealed that it harbors a potential YY1 binding sequence. Although we found no association between SPDEF methylation and its gene expression in our blood-based sample, several associations with trans-genes were evident. An enrichment analysis resulted in an overrepresentation of genes involved in opioid-related disorders, alcoholism, and tobacco use disorder. Genes such as OPRM1 and DRD2 have been extensively studied and found to influence the endogenous opioid system (31) and dopamine regulation (32), which are known to be involved in addiction (33). Furthermore, we found that the SPDEF promoter region also physically interacted with DRD2, suggesting an involvement of SPDEF in the regulation of DRD2 gene expression.
SPDEF is highly expressed in the prostate but is also expressed in the brain and liver, the two principal organs affected by alcohol abuse (34). Although we cannot rule out the possibility that the effect of SPDEF methylation on substance use is liver dependent, we found an interaction of SPDEF methylation and psychosocial stress with the right caudal cuneus gyrus. An inverse relationship between positron emission tomography-DRD2 receptor availability and functional MRI activation in the cuneus has been observed previously (35). It is tempting to speculate that stress-induced SPDEF methylation may influence dopaminergic neurotransmission and affect this relationship. Various studies have also reported that early visual processing areas, such as the cuneus, are affected by the reward history of stimuli (36). One possible interpretation is that the cuneus is more susceptible to value-based modulation in individuals with higher stress-induced SPDEF methylation, which may increase the risk of substance abuse. In addition, the occipital lobe, which includes the cuneus, has been consistently reported to be involved in stress- and anxiety-related conditions, especially in females (37, 38), but the neurophysiological correlates of these observations remain to be elucidated.
Lastly, we found that SPDEF methylation at site cg01395541 is allele-specific. In our sample, individuals with the SPDEF risk genotype (GG and GA) showed greater variability in DNA methylation, psychosocial stress exposure, and substance-related behaviors. This is consistent with the idea of differential susceptibility, whereby individuals with particular genetic factors may be more vulnerable to their environment (39). We found and replicated a gene-environment interaction of stress and SNP with SPDEF methylation, suggesting that adolescents who harbor the minor allele are more sensitive to psychosocial stress exposure and may have a higher risk of developing substance use behaviors compared with adolescents with an allele type incapable of becoming methylated (AA homozygotes).
Although complex traits are polygenic, a limitation of our study is that a single gene was identified. This was likely a result of noise limitations inherent in genome-wide analyses, which only allowed for SPDEF to reach statistical significance. However, the identification of SPDEF is important because it enabled us to distinguish potential novel mechanisms underlying the risk for substance use. We were unable to detect clear gender effects because of the limited sample size. It is therefore possible that we did not identify additional potential mechanisms related to the androgenic function of the gene. We were able to measure methylation and gene expression only in peripheral blood, although it has become increasingly clear that individual variance in methylation across tissues can be conserved (40). The high correlation between blood-based SPDEF methylation and brain tissues (r2>0.8) indicates that SPDEF can be viewed as a potential biomarker for risk of developing substance abuse (23). Yet these harmonized changes in methylation may not have synonymous effects on gene expression (41), allowing for the possibility of a cis effect on SPDEF expression in the brain.
In summary, our findings describe a novel epigenetic mechanism that helps explain how psychosocial stress exposure influences adolescent substance abuse, thus providing evidence of variations in DNA methylation mediating the effect of environmental influences on psychiatric symptoms and disorders.
1 : Environmental stress and psychiatric illness. Biomed Pharmacother 2000; 54:135–141Crossref, Medline, Google Scholar
2 : Why do many psychiatric disorders emerge during adolescence? Nat Rev Neurosci 2008; 9:947–957Crossref, Medline, Google Scholar
3 : The neurobiology of intervention and prevention in early adversity. Annu Rev Clin Psychol 2016; 12:331–357Crossref, Medline, Google Scholar
4 : Life stress in adolescence predicts early adult reward-related brain function and alcohol dependence. Soc Cogn Affect Neurosci 2015; 10:416–423Crossref, Medline, Google Scholar
5 : Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 2003; 33(suppl):245–254Crossref, Medline, Google Scholar
6 : Epigenetics of stress, addiction, and resilience: therapeutic implications. Mol Neurobiol 2016; 53:545–560Crossref, Medline, Google Scholar
7 : Annual Research Review: DNA methylation as a mediator in the association between risk exposure and child and adolescent psychopathology. J Child Psychol Psychiatry 2018; 59:303–322Crossref, Medline, Google Scholar
8 : Prodynorphin CpG-SNPs associated with alcohol dependence: elevated methylation in the brain of human alcoholics. Addict Biol 2011; 16:499–509Crossref, Medline, Google Scholar
9 : Epigenetic interactions between alcohol and cannabinergic effects: focus on histone modification and DNA methylation. J Alcohol Drug Depend 2017; 5:259Crossref, Medline, Google Scholar
10 : TLR4 methylation moderates the relationship between alcohol use severity and gray matter loss. J Stud Alcohol Drugs 2017; 78:696–705Crossref, Medline, Google Scholar
11 : Current and future prospects for epigenetic biomarkers of substance use disorders. Genes (Basel) 2015; 6:991–1022Crossref, Medline, Google Scholar
12 : Methylation of the leukocyte glucocorticoid receptor gene promoter in adults: associations with early adversity and depressive, anxiety and substance-use disorders. Transl Psychiatry 2016; 6:e848Crossref, Medline, Google Scholar
13 : The Psychiatric GWAS Consortium: big science comes to psychiatry. Neuron 2010; 68:182–186Crossref, Medline, Google Scholar
14 : The IMAGEN study: reinforcement-related behaviour in normal brain function and psychopathology. Mol Psychiatry 2010; 15:1128–1139Crossref, Medline, Google Scholar
15 : Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol 2012; 41:200–209Crossref, Medline, Google Scholar
16 : Analysing and interpreting DNA methylation data. Nat Rev Genet 2012; 13:705–719Crossref, Medline, Google Scholar
17 . A multidimensional assessment of stressful life events among adolescents: derivation and correlates. 1981:400–415Google Scholar
18 : The ESPAD Report: Alcohol and Other Drug Use Among Students in 35 European Countries. Stockholm, Modintryckoffset AB, 2003Google Scholar
19 Illumina: Infinium HumanMethylation450 BeadChip. San Diego, Illumina, 2012. https://www.illumina.com/content/dam/illumina-marketing/documents/products/datasheets/datasheet_humanmethylation450.pdfGoogle Scholar
20 IBM: IBM SPSS Statistics for Windows. Armonk, NY, IBM, 2011Google Scholar
21 : Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014; 30:1363–1369Crossref, Medline, Google Scholar
22 : Impact of SNPs on methylation readouts by Illumina Infinium HumanMethylation450 BeadChip Array: implications for comparative population studies. BMC Genomics 2015; 16:1003Crossref, Medline, Google Scholar
23 : Interindividual methylomic variation across blood, cortex, and cerebellum: implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics 2015; 10:1024–1032Crossref, Medline, Google Scholar
24 . Matrix eQTL: ultra fast eQTL analysis via large matrix operations. 2012; 28:1353–1358Google Scholar
25 : Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44–57Google Scholar
26 : The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Gonome Biol 2018; 19:151Crossref, Medline, Google Scholar
27 : Quantitative cross-validation and content analysis of the 450k DNA methylation array from Illumina, Inc. BMC Res Notes 2012; 5:210–210Crossref, Medline, Google Scholar
28 : GWAS identifies four novel eosinophilic esophagitis loci. Nat Commun 2014; 5:5593Crossref, Medline, Google Scholar
29 : Signatures of prostate-derived ETS factor (PDEF) in cancer. Tumour Biol 2016; 37:14335–14340Crossref, Medline, Google Scholar
30 GeneCards: The Human Gene Database v4.5.0 Build 25. 2017. http://www.genecards.org/Google Scholar
31 : OPRM1 SNP (A118G): involvement in disease development, treatment response, and animal models. Drug Alcohol Depend 2010; 108:172–182Crossref, Medline, Google Scholar
32 : Gene expression profiling in porcine maternal infanticide: a model for puerperal psychosis. Am J Med Genet B Neuropsychiatr Genet 2008; 147B:1126–1137Crossref, Medline, Google Scholar
33 : Is there a common molecular pathway for addiction? Nat Neurosci 2005; 8:1445–1449Crossref, Medline, Google Scholar
34 : Genetic polymorphism in ethanol metabolism: acetaldehyde contribution to alcohol abuse and alcoholism. Mol Psychiatry 2004; 9:570–581Crossref, Medline, Google Scholar
35 : Association between striatal dopamine D2/D3 receptors and brain activation during visual attention: effects of sleep deprivation. Transl Psychiatry 2016; 6:e828Crossref, Medline, Google Scholar
36 : Sensory and motor aspects of addiction. Behav Brain Res 2010; 207:215–222Crossref, Medline, Google Scholar
37 : Transdiagnostic brain responses to disorder-related threat across four psychiatric disorders. Psychol Med 2017; 47:730–743Crossref, Medline, Google Scholar
38 : Gender differences in neural correlates of stress-induced anxiety. J Neurosci Res 2017; 95:115–125Crossref, Medline, Google Scholar
39 : For better and for worse. Curr Dir Psychol Sci 2007; 16:300–304Crossref, Google Scholar
40 : Methylomic profiling across brain and blood: Brain area-specific differentially methylated regions, individual differences, and allele-specific DNA methylation, Presented at proceedings of the 12th International Congress of Human Genetics/61st Annual Meeting of the American Society of Human Genetics, October 12, 2011, MontrealGoogle Scholar
41 : CpG island methylation pattern in different human tissues and its correlation with gene expression. Biochem Biophys Res Commun 2009; 383:421–425Crossref, Medline, Google Scholar



