Impact of CYP2C19 Genotype on Escitalopram Exposure and Therapeutic Failure: A Retrospective Study Based on 2,087 Patients
Abstract
Objective:
The antidepressant escitalopram is predominantly metabolized by the polymorphic CYP2C19 enzyme. The authors investigated the effect of CYP2C19 genotype on exposure and therapeutic failure of escitalopram in a large patient population.
Method:
A total of 4,228 escitalopram serum concentration measurements from 2,087 CYP2C19-genotyped patients 10–30 hours after drug intake were collected retrospectively from the drug monitoring database at Diakonhjemmet Hospital in Oslo. The patients were divided into subgroups based on CYP2C19 genotype: those carrying inactive (CYP2C19Null) and gain-of-function (CYP2C19*17) variant alleles. The between-subgroup differences in escitalopram exposure (endpoint: dose-harmonized serum concentration) and therapeutic failure (endpoint: switching to another antidepressant within 1 year after the last escitalopram measurement) were evaluated by multivariate mixed model and chi-square analysis, respectively.
Results:
Compared with the CYP2C19*1/*1 group, escitalopram serum concentrations were significantly increased 3.3-fold in the CYP2C19Null/Null group, 1.6-fold in the CYP2C19*Null/*1 group, and 1.4-fold in the CYP2C19Null/*17 group, whereas escitalopram serum concentrations were significantly decreased by 10% in the CYP2C19*1/*17 group and 20% in the CYP1C19*17/*17 group. In comparison to the CYP2C19*1/*1 group, switches from escitalopram to another antidepressant within 1 year were 3.3, 1.6, and 3.0 times more frequent among the CYP2C19Null/Null, CYP2C19*1/*17, and CYP1C19*17/*17 groups, respectively.
Conclusions:
The CYP2C19 genotype had a substantial impact on exposure and therapeutic failure of escitalopram, as measured by switching of antidepressant therapy. The results support the potential clinical utility of CYP2C19 genotyping for individualization of escitalopram therapy.
Affective disorders, with depression leading the way, are a major contributor to the morbidity caused by diseases worldwide (1, 2). Selective serotonin reuptake inhibitors (SSRIs), despite their limitations, are the cornerstone of modern anxiolytic and antidepressant pharmacotherapy (3). According to previous clinical and in vitro reports, escitalopram is the most selective (4, 5) and is among the most efficient of SSRIs in the therapy of depression (6, 7). However, therapeutic failure caused by a lack of response or by side effects in a significant portion of treated patients limits the clinical usefulness of escitalopram (8). The high interindividual variation in escitalopram metabolism can mean either very high or very low escitalopram serum levels, potentially increasing the incidence of side effects or leading to insufficient clinical response and subsequently resulting in therapeutic failure (9).
The initial biotransformation of escitalopram is predominantly catalyzed by the hepatic cytochrome P450 (CYP) enzyme CYP2C19 (10, 11). The CYP2C19 gene is highly polymorphic, and its genotypes determine the CYP2C19 enzymatic capacity; CYP2C19*2 is the main loss-of-function (Null) allele (12), while CYP2C19*17 is thus far the only discovered gain-of-function allele (13). Over the past decade, many studies explored the effect of CYP2C19 genetic polymorphism on escitalopram exposure and treatment success. However, these studies have produced conflicting findings and failed to determine reliably whether CYP2C19 genotype is a clinically relevant feature in escitalopram therapy. Some studies were performed in controlled settings but did not include a sufficient number of patients and therefore did not have the power to determine or quantify the consequences of CYP2C19 variant alleles (14, 15). Other studies included more patients but were performed under flexible-dosing protocols and without escitalopram concentration monitoring (8, 16). In such settings, it is difficult to evaluate the effect of CYP2C19 polymorphism on the therapeutic success of escitalopram. Consequently, unequivocal evidence is lacking for the clinical utility of CYP2C19 genotyping in escitalopram-treated patients. Thus, in most psychiatric clinics worldwide, escitalopram therapy usually starts with the standard recommended dosage of 10 mg/day (9), regardless of CYP2C19 genotype.
In this study, in a population of 2,087 CYP2C19-genotyped escitalopram-treated patients for whom concentration monitoring data were available, we sought to determine the exact contribution of CYP2C19 polymorphism to escitalopram exposure and to evaluate the extent to which patients of different CYP2C19 genotypes were prone to switching from escitalopram to other antidepressants. The results indicate that CYP2C19 genotype has a substantial impact on escitalopram exposure and therapeutic failure, as measured by switching of drug therapy.
Method
Patients
Data from patients undergoing treatment with escitalopram were retrospectively drawn from the database of a therapeutic drug monitoring (TDM) service at Diakonhjemmet Hospital in Oslo. Inclusion of patients from the TDM database was carried out by matching information about individuals for whom CYP2C19 genotyping had been performed and serum concentrations of escitalopram measured as a part of the clinical follow-up. In addition to CYP2C19 genotype and escitalopram serum concentrations, data on daily dosing, time between last drug intake and blood sampling, age, and gender were drawn from the TDM database (Table 1). To obtain data on treatment history, longitudinal TDM profiles of all the included patients were reviewed to register possible use and monitoring of antidepressants other than escitalopram. The TDM assays at the laboratory included analyses of all antidepressants available for clinical use in Norway.
| Genotype | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Measure | Null/Null (N=88) | *1/Null (N=473) | *17/Null (N=121) | *1/*1 (N=837) | *1/*17 (N=507) | *17/*17 (N=97) | ||||||
| N | % | N | % | N | % | N | % | N | % | N | % | |
| Female | 60 | 68 | 313 | 66 | 73 | 60 | 508 | 61 | 312 | 62 | 56 | 58 |
| Number of serum samples | 204 | 917 | 196 | 1,621 | 1,082 | 207 | ||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Age (years) | 48.5 | 19.1 | 47.2 | 19.2 | 49.4 | 19.7 | 47.4 | 18.9 | 48.6 | 20.1 | 44.8 | 17.6 |
| Drug dosage (mg/day) | 12.6 | 6.0 | 15.8 | 9.3 | 14.5 | 6.3 | 18.0 | 11.4 | 18.1 | 9.8 | 17.1 | 9.7 |
| Time between drug intake and blood sampling (hours) | 20.8 | 5.5 | 21.5 | 5.2 | 21.5 | 5.0 | 21.7 | 5.0 | 21.8 | 5.0 | 22.3 | 4.6 |
TABLE 1. Characteristics of the Six CYP2C19 Genotype-Defined Study Subgroupsa
Escitalopram concentrations were determined by a fully validated ultra-performance liquid chromatography–tandem mass spectrometry (UPLC-MS/MS) method, and CYP2C19 genotype analyses were performed by TaqMan-based assays including the following variant alleles: CYP2C19*2 (rs4244285), CYP2C19*3 (rs4986893/rs57081121), CYP2C19*4 (rs28399504), and CYP2C19*17 (rs12248560). CYP2C19*2, *3, and *4 are together classified as loss-of-function variant alleles (CYP2C19Null), whereas CYP2C19*17 is the only known gain-of-function variant allele (www.pharmvar.org). The allelic frequency of CYP2C19Null (18.4%) and CYP2C19*17 (19.8%) alleles were equivalent to those observed in Europeans in general (17). The patients were categorized into CYP2C19 genotype-defined metabolizer subgroups. Poor metabolizers (CYP2C19Null/Null) lacked a functional CYP2C19 enzyme. Intermediate metabolizers (CYP2C19*1/Null and CYP2C19*17/Null) carried one loss-of-function allele. Patients with the CYP2C19Null/*17 genotype were categorized as intermediate metabolizers because the CYP2C19Null allele has a much greater impact on phenotype than the gain-of-function CYP2C19*17 allele. Extensive metabolizers (CYP2C19*1/*1) were defined as patients who were homozygous for the wild type CYP2C19*1 allele and represented the reference genotype. Ultrarapid metabolizers were defined as those carrying CYP2C19*1/*17 or CYP2C19*17/*17.
The study was approved by the Regional Committee for Medical and Health Research Ethics and the Hospital Investigational Review Board. Ethical approval was given without requirement of patient consent because the study was based on existing data retrospectively retrieved from a routine TDM service.
Study Endpoints and Statistical Analysis
All statistical analyses were performed with IBM SPSS Statistics for Windows, version 22.0 (IBM, Armonk, N.Y.).
Escitalopram exposure.
To determine the effect of CYP2C19 genotype on escitalopram exposure, the measured steady-state serum concentrations were compared between the CYP2C19 genotype–defined subgroups. Prior to the statistical analysis, escitalopram concentrations were normalized to the standard 10 mg/day dosage and natural log transformed. The natural logarithm of the normalized escitalopram serum levels (c) was used because of its linear dependence on the time between the drug intake and blood sampling (t) and the elimination rate constant (Ke) according to the equation Δln(c)=Ke × t. Only concentrations measured in serum samples drawn from 10 to 30 hours after the latest dose intake were included in the exposure-versus-genotype evaluation.
A multivariate linear mixed-model analysis was used to evaluate the impact of CYP2C19 genotype on dose-harmonized escitalopram concentration. This analysis allows repeated measurements and is regarded as the optimal statistical model when the number of measurements available for the same individual is variable. The analysis was conducted with escitalopram serum concentrations as the dependent variable and CYP2C19 genotype–defined groups as independent variables; age and sampling time were included as covariates. The estimated marginal mean of each genotype-defined group in the model was calculated and reported at the mean of the covariates—i.e., age 48 years and sampling time 21.7 hours—and compared with the mean calculated for the reference genotype.
Therapeutic failure.
A switch from escitalopram to another antidepressant within 1 year after the last TDM analysis of escitalopram was defined as the primary endpoint when evaluating whether the CYP2C19 genotype was associated with therapeutic failure of escitalopram. This analysis was performed by a longitudinal review of each patient’s TDM profile. If a TDM analysis of one or more antidepressants other than escitalopram had been performed during the 1-year follow-up period after the last TDM analysis of escitalopram, this was defined as an occurrence of therapeutic failure (regardless of cause). Switch to another antidepressant within this time frame most likely represents the same episode of depression and hence reflects a therapeutic failure of escitalopram, due to either insufficient clinical response or adverse effects.
When assessing therapeutic failure in relation to the CYP2C19 genotype, the odds for switching from escitalopram to another antidepressant were compared between patients carrying variant genotypes and the reference genotype (CYP2C19*1/*1) using the chi-square test. Odds ratios and 95% confidence intervals were calculated for each of the variant genotype subgroups.
Subtherapeutic concentration.
As a secondary endpoint evaluating the impact of genotype on risk of therapeutic failure of escitalopram, concentration measurements below a defined minimal therapeutic boundary (subtherapeutic concentrations) were applied. The lower boundary was defined using the experimentally determined serotonin reuptake (SERT) inhibition constant of 9.2 nM (18) in combination with data from a study showing that the CSF concentration is one-third that of serum (19). From this information, the minimal therapeutic concentration was defined as 25 nM, which is in concordance with less than 50% in vivo SERT binding measured by positron emission tomography in humans (20). A SERT binding of 60%−80% is regarded as necessary for SSRIs to be effective (20). Considering that 15 ng/mL (46 nM) is defined as the lower reference level of escitalopram in the Association for Neuropsychopharmacology and Pharmacopsychiatry’s Consensus Guidelines for Therapeutic Drug Monitoring in Psychiatry (21), the use of 25 nM as a boundary for defining subtherapeutic escitalopram serum concentrations is reasonable. For the evaluation of differences in the portion of the measurements below 25 nM between variant CYP2C19 genotype–defined subgroups, the chi-square test was applied with the CYP2C19*1/*1 reference subgroup as well.
Results
Escitalopram serum concentration was determined in 2,087 CYP2C19-genotyped patients 10 to 30 hours after drug intake. Figure 1A illustrates the dosage-harmonized serum concentrations of escitalopram according to the predicted order of CYP2C19 metabolic capacity, i.e., CYP2C19Null/Null, CYP2C19*1/Null, CYP2C19*17/Null, CYP2C19*1/*1, CYP2C19*1/*17, and CYP2C19*17/*17.
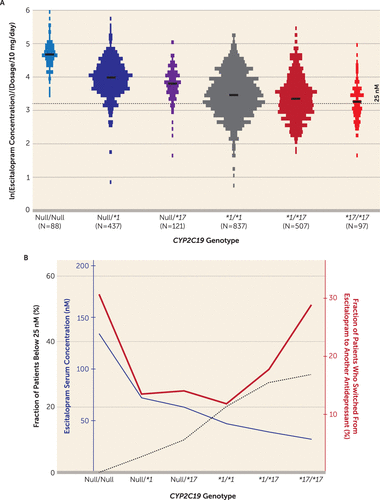
FIGURE 1. Effect of CYP2C19 Polymorphism on Escitalopram Serum Concentration and Therapeutic Failure Riska
a In panel A, escitalopram serum concentrations (measured 10–30 hours after the latest dose) are presented as a function of CYP2C19 genotype. The groups were defined by the presence of CYP2C19Null and CYP2C19*17 alleles and sorted according to the predicted increase of CYP2C19 metabolic capacity: poor metabolizers (CYP2C19Null/Null), intermediate metabolizers (CYP2C19*1/Null and CYP2C19*17/Null), extensive metabolizers (CYP2C19*1/*1), and ultrarapid metabolizers (CYP2C19*1/*17 and CYP2C19*17/*17). The escitalopram serum concentrations were normalized to the dosage of 10 mg/day and analyzed in the natural logarithmic scale. As predicted, compared with the extensive metabolizers, escitalopram serum levels were increased in the poor metabolizers and intermediate metabolizers and decreased in the ultrarapid metabolizers. In panel B, escitalopram serum concentration, the fraction of the patients whose serum concentration was below 25 nM, and the fraction of the patients who switched from escitalopram to another antidepressant are depicted as a function of CYP2C19 genotype (black, blue, and red lines). Conversely to the changes in the escitalopram concentration levels, compared with the extensive metabolizers, the fraction of the patients whose escitalopram serum concentrations were below the lower boundary of escitalopram’s therapeutic concentration range was increased in the ultrarapid metabolizers and decreased in the poor and intermediate metabolizers. Compared with the extensive metabolizers, the fraction of the patients who switched from escitalopram to another antidepressant formed a U-shaped curve, with increases in the ultrarapid and poor metabolizers. The data on effect sizes and statistical significance of the changes depicted in this figure are summarized in Table 2.
To account for the variable number of measurements per patient, the effect of CYP2C19 genotype on escitalopram concentration was estimated by a multivariate mixed-model analysis with age and sampling time as covariates. The serum concentration of escitalopram was significantly different between the CYP2C19 genotype-defined subgroups (Table 1). Mean estimates for escitalopram serum concentration were significantly greater among the poor and intermediate metabolizers compared with the extensive metabolizers—3.3 times greater for the poor metabolizers and 1.6 times (Null/*1) and 1.4 times (Null/*17) greater for the intermediate metabolizers (p<0.001) (Table 2). At the other end, mean escitalopram serum concentrations were significantly lower among the ultrarapid metabolizers, by approximately 10% (CYP2C19*1/*17) (p<0.003) and 20% (CYP2C19*17/*17) (p<0.002) compared with the extensive metabolizers (Table 2). In the mixed-model analysis, age and sampling time were found to be significant nongenetic covariates in explaining individual variability in escitalopram serum concentration. The dynamics of the increase in escitalopram exposure with age were similar regardless of CYP2C19 genotype; the concentration remained fairly constant in patients under age 50, while a pronounced increase was observed in patients over age 65 (Figure 2).
| Genotype | ||||||
|---|---|---|---|---|---|---|
| Measure | Null/Null (N=88) | Null/*1 (N=437) | Null/*17 (N=121) | *1/*1 (N=837) | *1/*17 (N=507) | *17/*17 (N=97) |
| Drug concentration (nM)/dosage (10 mg/day) | ||||||
| Concentration | 104.5 | 52 | 44.5 | 32 | 28.8 | 26.1 |
| 95% CI | 91.9–118.7 | 49.1–55.2 | 39.8–49.8 | 30.7–33.4 | 27.3–30.4 | 23.1–29.5 |
| Fold change | ↑3.270 | ↑1.627 | ↑1.392 | N/A | ↓0.899 | ↓0.817 |
| ln(c) | 4.649 | 3.952 | 3.796 | 3.465 | 3.36 | 3.263 |
| 95% CI | 4.521–4.777 | 3.894–4.011 | 3.684–3.908 | 3.423–3.508 | 3.306–3.414 | 3.141–3.385 |
| p | 2.4×10–62 | 5.6×10–39 | 5.8×10–8 | N/A | 0.003 | 0.002 |
| Serum drug levels below 25 nM | ||||||
| Frequency | 0 | 21 | 12 | 168 | 139 | 29 |
| % | 0 | 4.8 | 9.9 | 20.1 | 27.4 | 29.9 |
| p | 1.5×10–8 | 6.0×10–15 | 0.006 | N/A | 0.002 | 0.035 |
| Odds ratio | ↓0 | ↓0.201 | ↓0.438 | N/A | ↑1.504 | ↑1.698 |
| 95% CI | N/A | 0.126–0.322 | 0.236–0.815 | N/A | 1.162–1.947 | 1.065–2.708 |
| Switch to alternative antidepressant | ||||||
| Frequency | 27 | 59 | 17 | 99 | 90 | 28 |
| % | 30.7 | 13.5 | 14 | 11.8 | 17.8 | 28.9 |
| p | 1.0×10–5 | 0.42 | 0.46 | N/A | 0.003 | 2.7×10–5 |
| Odds ratio | ↑3.300 | 1.164 | 1.219 | N/A | ↑1.609 | ↑3.025 |
| 95% CI | 2.003–5.436 | 0.824–1.644 | 0.700–2.120 | N/A | 1.180–2.193 | 1.859–4.922 |
TABLE 2. Impact of CYP2C19 Genotype on Escitalopram Exposure and Therapeutic Failurea
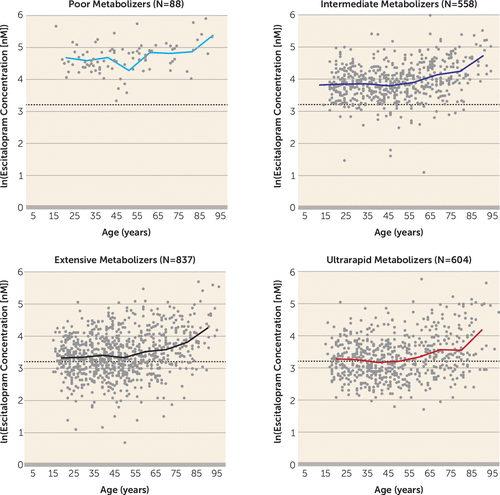
FIGURE 2. Effect of Age and Metabolizing Phenotypes on Escitalopram Serum Concentrationa
a Escitalopram serum concentrations (measured 10–30 hours after the latest dose) are presented as a function of age. Age did not significantly affect escitalopram serum concentration in patients under age 50, whereas an increase was observed in patients over age 65.
In the evaluation of individual measurements below the lower therapeutic boundary of escitalopram concentration defined for sufficient SERT inhibition (i.e., 25 nM), the proportion of patients with subtherapeutic levels increased with CYP2C19 metabolic capacity (Table 2). Compared with the reference group (patients homozygous for CYP2C19*1), which comprised 20.1% patients with subtherapeutic levels, the proportion of patients with escitalopram serum concentrations below 25 nM was significantly lower among the poor and intermediate metabolizers (p<0.001) and significantly higher among the ultrarapid metabolizers (p<0.003) (Table 2). While none of the poor metabolizers had escitalopram serum concentrations below 25 nM after the integration of multiple concentration measurements, the odds ratio estimates for the incidence of concentrations below this boundary were 0.20 (CYP2C19Null/*1) and 0.44 (CYP2C19Null/*17) for the intermediate metabolizer subgroups and 1.5 (CYP2C19*1/*17) and 1.7 (CYP2C19*17/*17) for the ultrarapid metabolizers compared with the extensive metabolizers (Table 2).
To address the effect of CYP2C19 genotype on the probability of escitalopram therapeutic failure in naturalistic settings, the fraction of patients who switched to another antidepressant within 1 year after the last TDM analysis of escitalopram was compared between the subgroups. In the extensive metabolizer reference group, the 1-year switch frequency was 11.8% (Table 2). Both the poor metabolizers and the ultrarapid metabolizers showed significantly higher frequencies of switching from escitalopram to another antidepressant compared with the extensive metabolizers, with odds ratios of 3.3 (p<0.001) for the poor metabolizers, 1.6 (p=0.003) for the CYP2C19*1/*17 subgroup, and 3.0 (p<0.001) for the CYP2C19*17/*17 subgroup. Regardless of CYP2C19 genotype, escitalopram was most often switched to venlafaxine (34% of the overall cases), a combined serotonin-norepinephrine reuptake inhibitor, followed by switch to another SSRI (25%).
The effect of CYP2C19 genotype on the various endpoints is depicted in Figure 1B, which illustrates the relationship between relative exposure and proportion with therapeutic failure, by subgroup. Interestingly, the fraction of patients who switched to another antidepressant within 1 year formed a U-shaped curve, with higher rates of drug switch among the patients with the lowest and highest metabolic capacity.
Discussion
The large number of patients included in this study allowed quantification of the effect of CYP2C19 genotype on escitalopram exposure and therapeutic failure, as monitored by drug switching, at a relatively high statistical power. The results provide strong evidence that CYP2C19 genotype has a major clinical impact on escitalopram therapy through a profound effect on escitalopram exposure. In particular, CYP2C19 genotype–defined ultrarapid metabolizers and poor metabolizers were linked with an elevated risk of therapeutic failure. These findings suggest that preemptive CYP2C19 genotyping could prospectively be of value for the individualization of escitalopram dosing, resulting in increased drug efficacy and fewer cases of drug switching.
According to the genotype-associated escitalopram concentrations, a high proportion of the ultrarapid metabolizers exhibited escitalopram serum levels below the defined lower limit of effective concentration at the standard dosing regimen (10 mg/day). This is the likely explanation for the higher chances of therapeutic failure in this subgroup, since the chronic effect of continuous lower levels of escitalopram, for weeks or months, causing insufficient receptor occupancy, will have a higher phenotypic effect than anticipated just from the serum levels. The necessity of achieving an adequate escitalopram serum concentration quickly is of particular relevance for certain patient subpopulations, such as those with high rates of nonadherence or with suicidal thoughts. According to our results, CYP2C19 genotype–defined ultrarapid metabolizers exhibit an increased incidence of therapeutic failure and exposure to subtherapeutic escitalopram serum concentrations. This may be related to our previous results (22) showing higher suicidality among suicide attempters who are ultrarapid CYP2C19 metabolizers as compared with poor metabolizers, intermediate metabolizers, or extensive metabolizers, based on a study of 207 depressed patients.
Poor metabolizers exhibited a 3.3-fold increase in drug concentrations compared with extensive metabolizers, caused by the absence of CYP2C19-mediated escitalopram metabolism. The effect on exposure was paralleled by a higher incidence of drug switching in poor metabolizers compared with extensive metabolizers. Well-replicated results have shown that the occurrence and severity of escitalopram-induced side effects such as insomnia, diarrhea, somnolence, dizziness, increased sweating, constipation, fatigue, and indigestion are dose dependent (23–26) and occur two to three times more frequently in patients receiving 20 mg/day compared with those receiving 10 mg/day. Therefore, the likely explanation for the higher frequency of therapeutic failure in poor metabolizers is an increased risk of dose-dependent side effects due to elevated escitalopram serum levels. Thus, the results from this study suggest that CYP2C19 genotype–guided dosing may have the potential to improve the clinical success of escitalopram therapy and could be considered as a possibility in routine clinical practice. Ultrarapid and poor metabolizers represent the subpopulations that would benefit the most from a successful individualization of escitalopram dosing based on CYP2C19 genotype. Ultrarapid and poor metabolizers together represent 33% of our study population. Considering that 16 million people in the United States (27) and more than 300 million people in the world (28) suffer from depression, preemptive genotyping for CYP2C19 could then be expected to increase the success rate of escitalopram-based antidepressant therapy for millions of patients around the world, and to a greater extent in countries with high prevalences of CYP2C19*2 and CYP2C19*17, notably, for example, in East and South Asia, where the frequency of CYP2C19*2 exceeds 30% (17).
The Clinical Pharmacogenetics Implementation Consortium (CPIC) (9) recommends 1) initiating therapy at the standard recommended starting dosage of escitalopram (10 mg/day) for intermediate metabolizers and extensive metabolizers; 2) considering either reducing the dosage of escitalopram by 50% or selecting an alternative drug not predominantly metabolized by CYP2C19 for poor metabolizers; and 3) selecting an alternative drug not predominantly metabolized by CYP2C19 for ultrarapid metabolizers. The data presented here suggest the need for further discussion on the escitalopram dosing recommendations. According to our exposure-by-genotype data in this study, it appears that 5 mg/day is generally sufficient to reach therapeutic serum levels in poor metabolizers, whereas for intermediate and extensive metabolizers, the standard dosage (10 mg/day) is more appropriate, as suggested by the CPIC guidelines. Young ultrarapid metabolizers appear to be a group for whom a dosage increase from 10 mg/day to 20 mg/day is frequently necessary to reach sufficient serum levels of escitalopram (Figure 1 and Figure 2). Of course, in certain cases, TDM can be utilized to support decisions on whether to further modify escitalopram dosing, and it is of particular importance in patients with hepatic or renal functional impairments and patients on polypharmacy. Escitalopram exposure increases in the older ages, unrelated to CYP2C19 genotype, an effect that is likely due to an age-dependent decrease in hepatic or renal perfusion. The data presented here suggest that patients over age 65 are not likely to benefit from dosages above 10 mg/day even if they are in the ultrarapid metabolizer subgroup, which is in agreement with the U.S. Food and Drug Administration drug label for escitalopram (29).
Several limitations of this study are worth mentioning. One is the unavailability of data that could be used to estimate the patients’ volumes of distribution. Since the serum concentration of a drug at a specific time point is inversely proportional to the volume of distribution, it is likely that including this information as a covariate would significantly improve the estimate of the CYP2C19 genotype effects on escitalopram serum concentration. Information about comedication with drugs that potentially create pharmacokinetic interactions is another factor of importance for the variability in serum concentration of escitalopram. Lack of data on comedication therefore represents a study limitation, in line with other nongenetic factors that may also affect drug concentrations (e.g., comorbidity).
Second, although the available information on switching to other antidepressants can be used as a clear indication of therapeutic failure, the exact information on diagnosis and treatment outcome at the time of blood sampling cannot be obtained. Consequently, a follow-up hypothesis-oriented clinical study in more controlled settings is needed to evaluate the extent of the benefit of using CYP2C19 genotype to tailor escitalopram therapy.
Finally, despite all the data indicating that preemptive CYP2C19 genotyping can be useful in escitalopram therapy, the cost-effectiveness of such an intervention, which is of relevance for routine usage and reimbursement, is beyond the scope of this report. A previous study demonstrated the cost-effectiveness of preemptive CYP2C19/CYP2D6 genotyping in antipsychotic therapy (30), and a similar study is needed to assess the cost-benefit ratio of CYP2C19 genotyping in escitalopram therapy.
1 : Burden of depressive disorders by country, sex, age, and year: findings from the global burden of disease study 2010. PLoS Med 2013; 10:e1001547Crossref, Medline, Google Scholar
2 : Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012; 380:2163–2196Crossref, Medline, Google Scholar
3 : Diagnostic and Statistical Manual of Mental Disorders, 5th ed (DSM-5). Washington, DC, American Psychiatric Association, 2013Crossref, Google Scholar
4 : Escitalopram, the most selective SSRI: in vitro data and in vivo binding studies using the new selective SERT ligand 3H-MADAM. Eur Neuropsychopharmacol 2002; 12(suppl 3):S209Crossref, Medline, Google Scholar
5 : Neurological soft signs in the clinical course of schizophrenia: results of a meta-analysis. Front Psychiatry 2014; 5:185Crossref, Medline, Google Scholar
6 : A comparative review of escitalopram, paroxetine, and sertraline: are they all alike? Int Clin Psychopharmacol 2014; 29:185–196Crossref, Medline, Google Scholar
7 : Comparative efficacy and acceptability of 12 new-generation antidepressants: a multiple-treatments meta-analysis. Lancet 2009; 373:746–758Crossref, Medline, Google Scholar
8 : CYP2C19 variation and citalopram response. Pharmacogenet Genomics 2011; 21:1–9Crossref, Medline, Google Scholar
9 : Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther 2015; 98:127–134Crossref, Medline, Google Scholar
10 : Citalopram and escitalopram plasma drug and metabolite concentrations: genome-wide associations. Br J Clin Pharmacol 2014; 78:373–383Crossref, Medline, Google Scholar
11 : The clinical pharmacokinetics of escitalopram. Clin Pharmacokinet 2007; 46:281–290Crossref, Medline, Google Scholar
12 : Genetic tests which identify the principal defects in CYP2C19 responsible for the polymorphism in mephenytoin metabolism. Methods Enzymol 1996; 272:210–218Crossref, Medline, Google Scholar
13 : A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther 2006; 79:103–113Crossref, Medline, Google Scholar
14 : Kinetics of omeprazole and escitalopram in relation to the CYP2C19*17 allele in healthy subjects. Eur J Clin Pharmacol 2008; 64:1175–1179Crossref, Medline, Google Scholar
15 : Impact of the ultrarapid CYP2C19*17 allele on serum concentration of escitalopram in psychiatric patients. Clin Pharmacol Ther 2008; 83:322–327Crossref, Medline, Google Scholar
16 : Treatment outcomes of depression: the Pharmacogenomic Research Network Antidepressant Medication Pharmacogenomic Study. J Clin Psychopharmacol 2014; 34:313–317Crossref, Medline, Google Scholar
17 : Worldwide distribution of cytochrome P450 Alleles: a meta-analysis of population-scale sequencing projects. Clin Pharmacol Ther 2017; 102:688–700Crossref, Medline, Google Scholar
18 : The two enantiomers of citalopram bind to the human serotonin transporter in reversed orientations. J Am Chem Soc 2010; 132:1311–1322Crossref, Medline, Google Scholar
19 : Measuring citalopram in blood and central nervous system: revealing a distribution pattern that differs from other antidepressants. Int Clin Psychopharmacol 2016; 31:119–126Crossref, Medline, Google Scholar
20 : Therapeutic plasma concentrations of antidepressants and antipsychotics: lessons from PET imaging. Pharmacopsychiatry 2011; 44:236–248Crossref, Medline, Google Scholar
21 : AGNP consensus guidelines for therapeutic drug monitoring in psychiatry: update 2011. Pharmacopsychiatry 2011; 44:195–235Crossref, Google Scholar
22 : Elevated CYP2C19 expression is associated with depressive symptoms and hippocampal homeostasis impairment. Mol Psychiatry 2017; 22:1155–1163Crossref, Medline, Google Scholar
23 : Efficacy and tolerability of escitalopram in 12- and 24-week treatment of social anxiety disorder: randomised, double-blind, placebo-controlled, fixed-dose study. Depress Anxiety 2004; 19:241–248Crossref, Medline, Google Scholar
24 : Fixed-dose trial of the single isomer SSRI escitalopram in depressed outpatients. J Clin Psychiatry 2002; 63:331–336Crossref, Medline, Google Scholar
25 Forest Pharmaceuticals: Lexapro package insert. 2002. https://www.accessdata.fda.gov/drugsatfda_docs/label/2003/21323se1-003,se8-007,21365se8-001,se1-004_lexapro_lbl.pdfGoogle Scholar
26 US Food and Drug Administration: FDA Drug Safety Communication: Revised recommendations for Celexa (citalopram hydrobromide) related to a potential risk of abnormal heart rhythms with high doses. https://www.fda.gov/Drugs/DrugSafety/ucm297391.htmGoogle Scholar
27 National Institute of Mental Health: Major depression among adults. https://www.nimh.nih.gov/health/statistics/prevalence/major-depression-among-adults.shtmlGoogle Scholar
28 World Health Organization: Depression fact sheet. http://www.who.int/mediacentre/factsheets/fs369/en/Google Scholar
29 Forest Pharmaceuticals: Lexapro package insert. 2004. https://www.fda.gov/ohrms/dockets/ac/04/briefing/2004-4065b1-22-tab11C-Lexapro-Tabs-SLR015.pdfGoogle Scholar
30 : Does pharmacogenetic testing for CYP450 2D6 and 2C19 among patients with diagnoses within the schizophrenic spectrum reduce treatment costs? Basic Clin Pharmacol Toxicol 2013; 113:266–272Crossref, Medline, Google Scholar



