Revisiting Antipsychotic Drug Actions Through Gene Networks Associated With Schizophrenia
Abstract
Objective:
Antipsychotic drugs were incidentally discovered in the 1950s, but their mechanisms of action are still not understood. Better understanding of schizophrenia pathogenesis could shed light on actions of current drugs and reveal novel “druggable” pathways for unmet therapeutic needs. Recent genome-wide association studies offer unprecedented opportunities to characterize disease gene networks and uncover drug-disease relationships. Polygenic overlap between schizophrenia risk genes and antipsychotic drug targets has been demonstrated, but specific genes and pathways constituting this overlap are undetermined. Risk genes of polygenic disorders do not operate in isolation but in combination with other genes through protein-protein interactions among gene product.
Method:
The protein interactome was used to map antipsychotic drug targets (N=88) to networks of schizophrenia risk genes (N=328).
Results:
Schizophrenia risk genes were significantly localized in the interactome, forming a distinct disease module. Core genes of the module were enriched for genes involved in developmental biology and cognition, which may have a central role in schizophrenia etiology. Antipsychotic drug targets overlapped with the core disease module and comprised multiple pathways beyond dopamine. Some important risk genes like CHRN, PCDH, and HCN families were not connected to existing antipsychotics but may be suitable targets for novel drugs or drug repurposing opportunities to treat other aspects of schizophrenia, such as cognitive or negative symptoms.
Conclusions:
The network medicine approach provides a platform to collate information of disease genetics and drug-gene interactions to shift focus from development of antipsychotics to multitarget antischizophrenia drugs. This approach is transferable to other diseases.
Antipsychotics were incidentally discovered more than 60 years ago, but their mechanisms of action have still not been fully revealed. Antipsychotics are the main medication available to patients with schizophrenia but have little effect on negative and cognitive symptoms of the disease. Current antipsychotics are also limited by serious side effects (1, 2) that reduce treatment compliance (3), and about one-third of patients with psychosis are classified as treatment resistant (4). Thus, there is great need for improved medications for these patients, but drug development has been hampered by poor knowledge of disease etiology and underlying genetics. There is a growing interest in harnessing knowledge of risk genes for developing better treatments for common diseases (5–7). For example, Nelson et al. (8) found that drugs with genetically supported mechanisms of action succeeded in moving from phase 1 trials to gaining approval twice as often as drugs without genetic support. Schizophrenia is highly heritable (9), and a well-powered genome-wide association study (GWAS) identified as many as 108 independent loci related to the disease, containing more than 300 genes (10). Improved knowledge of these risk genes may be used to inform drug development by revealing potential mechanisms of action of current drugs and by identifying new drug targets. Antipsychotics bind to numerous proteins (11), of which dopamine and serotonin receptors are the only ones with known biological links to schizophrenia. However, a recent study found genetic overlap between schizophrenia risk genes and antipsychotic target genes, which suggests the pharmacological mechanisms may be polygenic and may also involve pathways that are not yet identified (12).
Most genotype-phenotype relationships arise from complexity of cellular interactions (13). Risk genes of polygenic disorders do not operate in isolation but in combination and interaction with other risk genes. The effect of a perturbation in one gene can propagate to affect other nearby proteins in the protein-protein interaction network, referred to here as the interactome. Therefore, protein products of genes that are associated with a particular disease tend to interact with one another and converge on related biological and functional networks (the so-called disease module) rather than being randomly spread throughout the interactome (14, 15). Thus, network biology (16) or network medicine (15) provides an important framework where knowledge of protein-protein interactions can be used to gain more comprehensive insight into the molecular mechanisms of complex diseases (14, 17).
The network approach also provides a unique opportunity to study drug effects by integrating the human interactome with knowledge of drug targets (18). The Drug-Gene Interaction Database (DGIdb) (11, 19) contains information on genes whose products are known to interact with drugs in humans (drug target genes), as well as genes that belong to the “druggable” genome (11, 20). Okada et al. (21) examined how targets of rheumatoid arthritis drugs map to the interactome neighborhood of identified risk genes for rheumatoid arthritis, and they found an overlap between drug target genes and risk genes as well as interactome neighbors of risk genes (21). Another study examined how drug target genes overlap with GWAS hits across a variety of diseases, and it found very little direct overlap (22). However, drug target genes showed threefold enrichment among the closest interactome neighbors, and enrichment was also significant among the second neighbors, suggesting that neighboring genes in the interactome should be included when searching for suitable candidates for drug repurposing (22).
Here, adopting a novel network biology approach (14), we study the schizophrenia disease module and its intersection with current antipsychotic drug target genes integrating information from the human interactome (14) with data from the DGIdb. Our objective was to inform development of new medications (20) by improving understanding of disease etiology and the function of current medications, as well as by identifying genes worth further examination as new drug targets or drug repurposing opportunities. We first identified and characterized a schizophrenia disease module in the interactome, and then we examined the interactome link between antipsychotic drug targets and schizophrenia risk genes through protein interactions. We revealed the specific risk genes and pathways that are involved in this link as well as the risk genes that are not linked to current antipsychotics. This information may be useful for identifying targets of future drugs that may also treat symptoms of schizophrenia other than psychosis.
Method
Interactome Database
The protein-protein interaction network used in our main analyses is the human interactome created by Menche et al. (14), consisting of 13,460 proteins interconnected by 141,296 high-confident physical interactions with experimental support. For comparison, we also used the STRING database (23) (see the data supplement that accompanies the online edition of this article).
Network Terminology and Illustrations
In the network terminology used throughout this article, nodes refer to genes or their corresponding gene products (proteins), and edges refer to connections between two genes through identified protein-protein interactions between their products. The distance (ds) between two nodes in the network is defined as the smallest number of edges, or steps, connecting them (i.e., shortest path length). Network figures were created using Cytoscape (24), with nodes illustrated as a shape (octagons for drug target genes, and circles for disease genes and neighbors of disease genes or drug targets) and edges as lines.
Disease Risk Genes
Schizophrenia risk genes were derived from the largest multicenter GWAS of schizophrenia to date (10) (36,989 cases and 113,075 controls), performed by the Psychiatric Genomics Consortium. In this GWAS, 108 independent loci were identified with a p value threshold of 5×10−8, which contained 326 potential causal protein coding genes (see Supplementary Table 3 in Ripke et al. [10]). In addition, one of the loci spans a large region on chromosome 6 containing the major histocompatibility complex (MHC). Because this locus contains hundreds of genes, all could not be included in the analyses as the majority might not be functionally related to schizophrenia. A recent article showed that the MHC-schizophrenia association is partly driven by the genes C4A and C4B (25). We therefore included these two genes in the analyses to represent the signal from the MHC region, resulting in a total of 328 risk genes included in the analyses.
Network Localization
To estimate network localization of the schizophrenia risk genes, we first calculated the shortest distance, ds, between each disease gene to the next closest disease gene and the corresponding frequency distribution, P(ds). To estimate if the disease genes were more localized than that expected by chance, 1,000 random sets of genes with the same number of genes as the disease set were generated to yield the distribution Prand(ds) from which test statistics could be calculated for the observed value of ds (see Menche et al. [14] for details). We also calculated the largest connected component of disease genes in the interactome space, containing S disease genes. S was compared with 1,000 random gene sets with the same number of genes to yield the distribution Prand(S). Following Menche et al., we define the schizophrenia disease module based on both ds and S. A statistically significant ds is driven by risk genes with the shortest distance to another risk gene (i.e., interconnected risk genes [d=1]), and genes in the largest connected component, S, are a subset of interconnected risk genes. Thus, we refer to interconnected risk genes as core genes of the disease module, or as the core disease module. We also performed control analyses with calculation of ds that takes node degree into account for the selection of random gene sets (26). Node degree was also incorporated to an alternative network localization method based on network neighborhood overlap (27) (see the online data supplement).
Antipsychotic Drug Targets
We included 64 drugs listed as antipsychotics in the Anatomical Therapeutic Chemical Classification System of the World Health Organization’s Collaborating Center for Drug Statistics Methodology. To identify drug-gene interactions in the interactome, we used data from the drug-gene interaction database (DGIdb version 2.22, http://dgidb.genome.wustl.edu, downloaded on Oct. 17, 2016), a freely accessible database for identifying known and potential drug-gene interactions (11, 19). We identified 88 genes targeted by at least one of the 64 antipsychotics. Eighty of these were included in the Menche et al. interactome with at least one interactor.
Gene Set Enrichment Analysis
We used MAGENTA (Meta-Analysis Gene-set Enrichment of variaNT Associations) (28) to test if antipsychotic drug targets, or their first interactome neighbors, were enriched for associations with schizophrenia compared with a large number of randomly selected genes (see the online data supplement for details). We also examined, as control analyses, potential enrichment of antipsychotic drug target genes among GWAS-identified risk genes for three control conditions: depressive symptoms (29), Alzheimer’s disease (30), and type 2 diabetes (31).
Network Graph of Genetic Overlap Between Antipsychotic Targets and Disease Genes
We calculated the number of interaction steps (edges) between each specific schizophrenia risk gene and its closest antipsychotic drug target in the interactome, with 0, 1, 2, or >2 steps.
Gene Ontology Enrichment
ToppGene (32) was used to examine enrichment in gene ontology annotations and pathways for the following subsets of risk gene sets related to the schizophrenia disease module: 1) all risk genes, 2) interconnected risk genes, 3) risk genes belonging to the largest connected component of risk genes (see Supplementary Table 1 in the data supplement). We also examined risk gene sets according to their shortest path length (distance) to antipsychotic drug targets in the interactome (0 or 1, 2, or >2 steps to an antipsychotic drug target; see Supplementary Table 2 in the data supplement). ToppGene uses hypergeometric distribution with Bonferroni correction for determining statistical significance. We included the gene ontology annotation categories molecular function and biological process, as well as pathways, with a Bonferroni-corrected p value threshold of 0.05.
Results
The Schizophrenia Disease Module
Using the Menche et al. interactome, 251 schizophrenia risk genes were included. Schizophrenia risk genes were significantly localized in the interactome compared with random gene sets (p=0.0015), with an average of 1.7 edges between two risk genes, forming a disease module (14). Eighty-two of 251 risk genes (33%) directly connected with at least one other risk gene (i.e., interconnected risk genes), as indicated by the outer circle in Figure 1. Three interconnected risk genes, CHRNA3, CHRNA5, and CHRNB4, were derived from the same loci in the GWAS, leaving 79 interconnected risk genes for gene ontology enrichment analyses. The largest connected component consisted of 32 risk genes (z=1.1, p=0.27), as indicated by the inner circle in Figure 1. Using the STRING (23) interactome, 254 schizophrenia risk genes were included. The shortest average distance between gene pairs was 1.59 edges, again significantly shorter than that for the random gene sets (p=5.2×10−4). The largest connected component consisted of 57 genes (z=2.2, p=0.028). Control analyses taking node degree into account (26) confirmed the main results of schizophrenia risk genes being significantly localized, both using the Menche et al. interactome (p=0.033) and the STRING database (p=0.006), as well as using the network neighborhood overlap method (see the online data supplement).
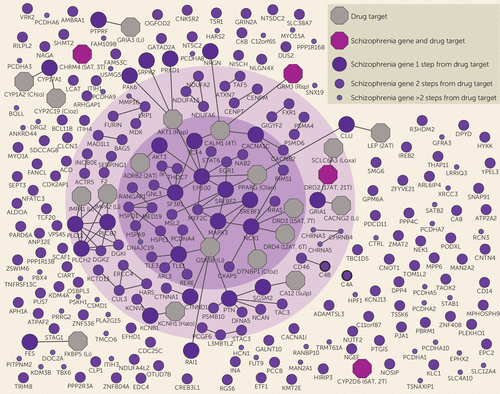
FIGURE 1. Connection Between Schizophrenia Risk Genes and Antipsychotic Drug Targetsa
a Schizophrenia risk genes are labeled by color and size according to their shortest path lengths to antipsychotic drug targets. Antipsychotic drug targets are shown as octagons, with the number of atypical (AT) and typical (T) antipsychotics in parentheses. Drug targets that do not interact with any risk gene are not shown. Abbreviated drug names are given for proteins that are targeted by a single antipsychotic: Chlo=chlorpromazine; Cloz=clozapine; Li=lithium; Loxa=loxapine; Olan=olanzapine; Risp=risperidone; Sulp=sulpiride. The inner circle indicates the genes belonging to the largest connected component, and the outer circle indicates the interconnected risk genes. We refer to these two gene set components as the core disease module, given that they are used as network properties to quantify the degree to which disease risk genes cluster in specific interactome neighborhoods. Risk genes close to antipsychotic drug targets were significantly overrepresented among interconnected risk genes (p=0.00023; chi-square test) (counts shown in Supplementary Table 4 in the online data supplement). For full details of specific antipsychotic targets, see Supplementary Table 3 in the data supplement. Further details on C4A and C4B (marked with thicker borders) are shown in Supplementary Figure 2 in the data supplement.
To gain further insight into the biological function of risk genes, we performed gene ontology and pathway enrichment analyses for a) all schizophrenia risk genes (N=328), b) interconnected risk genes (N=79), and c) risk genes belonging to the largest connected component in the interactome (N=32). All significant results from analyses a–c are presented in full detail in Supplementary Table 1 in the data supplement (p<0.05, corrected). Importantly, by restricting the analyses to genes in b) and c), the core genes of the disease module only, we were able to identify enrichment for several biological processes and pathways that could not be identified when treating all risk genes equally (33), including developmental biology, learning or memory, and cognition (Figure 2; see also Supplementary Table 1 in the data supplement).
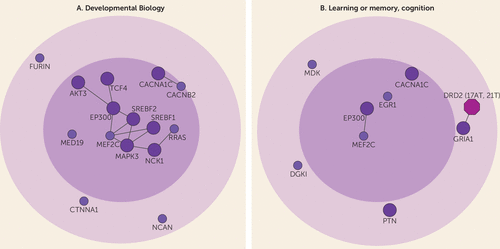
FIGURE 2. Significant Biological Processes and Pathways for the Schizophrenia Disease Modulea
a The figure shows two of the top ranked biological processes and pathways derived from gene ontology and pathway enrichment analysis for the interconnected risk genes (N=79). The developmental biology pathway (477129, REACTOME, p=6.3×10−9) is shown in panel A. The learning or memory (gene ontology:0007611, p=1.87×10−6) and cognition (gene ontology:0050890, p=4.64×10−6) pathway, which consisted of the same set of risk genes, is shown in panel B. See also Table 1 as well as Supplementary Table 1 in the online data supplement.
Examining the Link Between Antipsychotic Drug Targets and Schizophrenia
First, using gene set enrichment analysis (MAGENTA [28]), we found that antipsychotic drug target genes (N=88 from the DGIdb) were significantly enriched for associations with schizophrenia (p=0.0099) compared with random gene sets, confirming a previous report of polygenic overlap between schizophrenia risk genes and antipsychotic drug targets (12). Beyond replicating the previous finding, we incorporated network information because it has been shown that drug targets were substantially enriched in the nearest neighbors of GWAS genes (22). We identified 1,133 first neighbors of an antipsychotic drug target (proteins in direct protein-protein interaction with drug targets, excluding the 88 direct drug target genes) through the Menche et al. interactome and found that these were also significantly enriched for association with schizophrenia (p=0.0057). Twenty-seven of the 251 schizophrenia risk genes were first neighbors of drug target genes (see Supplementary Table 2A in the data supplement). No antipsychotic gene set enrichment was seen for the control conditions depressive symptoms (29), Alzheimer’s disease (30), or type 2 diabetes (31) (all p values >0.1).
To identify the specific risk genes and pathways that constitute the observed drug-disease link, we calculated the number of interactions between each risk gene and its closest antipsychotic drug target (Figure 1; see also Supplementary Table 2A in the data supplement). Four genes were both a risk gene and a drug target gene: glutamate metabotropic receptor 3 (GRM3), dopamine receptor D2 (DRD2), cholinergic receptor muscarinic 4 (CHRM4), and cytochrome P450 family 2 subfamily D member 6 (CYP2D6). Risk genes that overlap or connect with antipsychotic drug targets (≤1 step from drug targets) were overrepresented among interconnected risk genes as well as genes in the largest connected component (p=9.0×10−8 and p=0.00036, respectively; see also Supplementary Table 4 in the data supplement). For Alzheimer’s disease, used as a control condition, interconnected risk genes were not overrepresented among risk genes ≤1 step from antipsychotic drug targets (p=0.35). Gene ontology and pathway enrichment results of schizophrenia risk genes ≤1 step from drug targets revealed partly overlapping results to those of interconnected risk genes, such as learning or memory, and developmental biology (Table 1). In addition, enrichment was also seen for cholinergic, glutamatergic, and dopaminergic pathways (see Supplementary Table 2B in the data supplement). None of the schizophrenia risk genes that were not included in the interactome (N=77) were directly targeted by an antipsychotic drug, which is not extreme in comparison to all genes (p=0.89; see the data supplement for methods). For comparisons with the STRING database, see the supplementary methods and Supplementary Table 3 in the data supplement.
| Identification | Name | p |
|---|---|---|
| Biological processes | ||
| Gene ontology:0098916 | Anterograde trans-synaptic signaling | 8.05×10−8 |
| Gene ontology:0007611 | Learning or memory | 2.41×10−7 |
| Gene ontology:0051130 | Positive regulation of cellular component organization | 3.54×10−7 |
| Gene ontology:0010628 | Positive regulation of gene expression | 5.66×10−7 |
| Gene ontology:0050769 | Positive regulation of neurogenesis | 6.56×10−7 |
| Pathways | ||
| 477129 (REACTOME) | Developmental biology | 4.40×10−8 |
| 83085 (KEGG) | Long-term potentiation | 6.60×10−7 |
| 217716 (KEGG) | Cholinergic synapse | 8.84×10−6 |
| 213818 (KEGG) | Glutamatergic synapse | 1.01×10−5 |
| 868086 (KEGG) | Rap1 signaling pathway | 1.35×10−5 |
TABLE 1. Biological Processes and Pathways Related to Schizophrenia Risk Genes Overlapping With Current Drug Targetsa
Identification of Schizophrenia Risk Genes Untargeted by Antipsychotic Drugs
We next sought to map risk genes that were not connected to current antipsychotics (>2 steps, N=53; Figure 1; see also Supplementary Table 2 in the data supplement) together with their first interactome neighbors (N=122; see Supplementary Figure 1 in the data supplement). The purpose of this analysis is to identify druggable genes among molecular pathways involved in other aspects of schizophrenia than those targeted by antipsychotics as potential candidates for future drug development or repurposing. These genes were enriched for calcium ion binding and acetylcholine receptor–related molecular functions and pathways (see Supplementary Table 2D in the data supplement). Three genetic clusters from this map with potential as targets for cognitive enhancers in schizophrenia are shown in Figure 3. Risk genes with an intermediate link to drug targets (2 steps) showed few gene ontology enrichments (see Supplementary Table 2C in the data supplement). Finally, we examined if any existing drugs listed in the drug bank mapped onto schizophrenia risk genes >2 steps from an antipsychotic drug target, and we identified 11 approved and eight experimental drugs, as listed in Supplementary Table 5 in the data supplement.
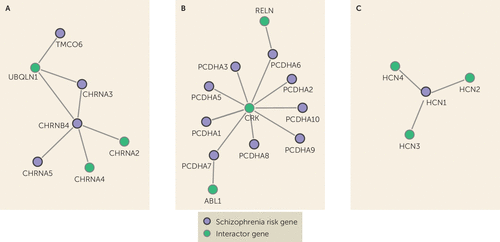
FIGURE 3. Schizophrenia Risk Gene Networks Untargeted by Current Antipsychoticsa
a Highlighted schizophrenia risk genes are shown with >2 steps or edges to antipsychotic drug target genes, with their first interactome neighbors. Panel A depicts the nicotinic acetylcholine receptor genes CHRNA3, CHRNA5, and CHRNB4. Panel B depicts Protocadherin alpha cluster genes PCDHA1–PCDHA10. Panel C depicts HCN1. We show three examples here; see Supplementary Figure 1 in the online data supplement for the full network.
Discussion
We examined the interactome link between antipsychotic drug targets and schizophrenia risk genes identified from the latest schizophrenia GWAS (10). First, we identified a schizophrenia disease module, characterized by core genes involved in developmental biology and cognition. Second, we found antipsychotic drug target genes, as well as their first interactome neighbors, to be enriched for association with schizophrenia. Through network graphs, we found the observed interactome link between existing drugs and the disease to be located among core genes in the disease module and to involve multiple pathways. Important risk genes that were not linked to current drug targets were also identified.
Using network methods and a high-confident human interactome (14), we showed that schizophrenia risk genes are significantly localized rather than randomly scattered in the interactome, forming a distinct disease module (14). Previous pathway analyses performed on schizophrenia risk genes from the latest large-scale GWAS (10) treated all risk genes equally and mainly identified enrichment for synapse- and dendrite-related pathways (33). One problem with the previous approach is that many GWAS-identified genetic loci contain several genes, of which all may not be causal to the disease. Importantly, it has been shown that when several genes are present in the same loci of a GWAS, risk genes that interact with other risk genes are more likely to be causally linked to the disease (34, 35). Restricting analyses to interconnected risk genes enabled the identification of novel and highly disease-relevant biological processes and pathways (see Supplementary Table 1B and 1C in the online data supplement). These core genes were most strongly related to developmental biology, which may constitute a central role in disease etiology (Figure 2A). Interestingly, core genes were also enriched for cognition-related processes and pathways (Figure 2B). Although schizophrenia is diagnosed based on positive and negative symptoms, cognitive impairment is a core clinical feature of the disease, with evidence of premorbid cognitive impairment from as young as age 7 (36).
We confirmed previously reported enrichment of antipsychotic drug target genes for association with schizophrenia (12) and, importantly, also found enrichment for neighbors of antipsychotic drug target genes. These results indicate that the pharmacological mechanisms of current antipsychotics overlap with the pathogenesis of schizophrenia and that the drug-disease link extends beyond dopamine to also involve other neurotransmitter systems, such as glutamatergic and cholinergic pathways (Table 1; see also Supplementary Table 2B in the data supplement). These pathways not only include risk genes directly targeted by antipsychotics (the glutamatergic and cholinergic receptor genes GRM3 and CHRM4) but also risk genes interacting with drug targets, such as CACNA1C, coding for a calcium channel subunit, and cell signaling kinases such as MAP kinase 3 (MAPK3) and AKT serine/threonine kinase 3 (AKT3). Indeed, previous efforts have identified potential drug targets within the cholinergic and glutamatergic systems (37), and pharmacogenetic studies have linked antipsychotic response to variants in GRM3 (38). Genetic variations in CYP2D6, a gene linked to drug metabolism and one of the schizophrenia risk genes directly targeted by antipsychotics, have been linked to side effects of antipsychotics (39). Some studies also linked variants in CYP2D6 to antipsychotic response, but results have been mixed (40). Although we did not observe significant overlap between antipsychotic targets and control conditions in our enrichment analyses, it remains possible for partial overlap at the individual gene level (e.g., pleiotropic variants), and this warrants future investigation.
Although genes close to current antipsychotics were related to learning and cognition, antipsychotics show only weak improvements of cognition in randomized controlled studies (41). Most antipsychotics reduce dopamine release, although the differential direction of altered dopamine levels in different brain areas has been linked to different aspects of schizophrenia (42–44). Dopamine dysfunction cannot explain all aspects of schizophrenia, and to develop improved medication, it is important also to focus on other potential targets. We identified the risk genes that are least connected to current antipsychotics together with their first neighbors (see Supplementary Figure 1 in the data supplement). This information may be used together with other sources of knowledge, such as the druggable genome (11), to inform future studies aiming to identify candidate targets for new drugs or drug repurposing to treat symptoms that are little affected by current antipsychotic medication, such as cognitive-enhancing drugs. Among these genes, important examples are nicotinic acetylcholine receptor genes (nAChRs) (Figure 3A). The nAChRs are thought to be important for the cognitive symptoms of schizophrenia (45) and are also implicated in Alzheimer’s disease (46, 47). The nAChRs are targeted by several existing drugs, including the Alzheimer’s disease drug galantamine (see the data supplement). Galantamine has been studied in humans with schizophrenia as well as in animal models, with indications of improved effect on cognitive symptoms and enhanced efficacy of antipsychotics in rats (48, 49). The nAChRs are also currently being examined as potential drug targets to enhance cognition in schizophrenia patients (50), further justifying our hypothesis that suitable drug targets may be found among these genes. Other interesting genes that did not overlap with antipsychotic drug genes are the Protocadherin gene cluster (PCDHA1–10) and hyperpolarization activated cyclic nucleotide gated potassium channel 1 (HCN1) (Figure 3B and 3C). The PCDHA genes are neural cell adhesion proteins involved in neural differentiation during development (51), processes thought to be important for the development of schizophrenia. Protocadherin genes have also been linked to cognition, personality, and mood disorders (52). The PCDHA genes are not considered druggable (11), but two neighbor genes are (RELN and ABL1). HCN1 is druggable (11) and may have a link to cognition in schizophrenia through involvement in mechanisms of synaptic plasticity and memory (53). Furthermore, these ion channels have been suggested as potential new targets for depression (54) and cognitive dysfunction in neurofibromatosis type 1 (55).
Limitations
A major limitation of the human interactome is potential bias toward well-studied proteins. To minimize this bias, the Menche et al. interactome also includes protein-protein interactions derived from unbiased high-throughput data sets (56–58), and we found that schizophrenia risk genes were significantly localized also when using node degree–preserved methods. Also, the known human protein-protein interaction network covers only an estimated 20% of all potential protein-protein interactions (14). However, the current level of network completeness was shown to successfully identify the disease module of 226 complex diseases (14). The available drug target data are also incomplete. Among the identified risk genes with more than two steps to a drug gene target, some may have undiscovered connections with drug target genes. To further address this limitation, we repeated our analyses using the less conservative STRING interactome, with consistent results.
Conclusions
By applying a network biology approach, we first identified a schizophrenia disease module, consisting of genes involved in developmental biology and cognition. Second, we found an overlap between the pathological mechanisms of schizophrenia and the pharmacological mechanisms of antipsychotics. This overlap involved protein interactions among drug targets and risk genes, which strengthens the proposal that network biology can be used to inform identification of new drug targets (59). Results might be used to advance the field of network pharmacology (59) in schizophrenia, with the potential development of more efficient multitarget drugs targeting not only psychosis but also other, poorly treated aspects of schizophrenia.
1 : Schizophrenia. N Engl J Med 2003; 349:1738–1749Crossref, Medline, Google Scholar
2 : Metabolic side effects of antipsychotic medication. Int J Clin Pract 2007; 61:1356–1370Crossref, Medline, Google Scholar
3 : Second-generation antipsychotics and extrapyramidal adverse effects. BioMed Res Int 2014; 2014:656370Crossref, Medline, Google Scholar
4 : Treatment-resistant schizophrenia: the role of clozapine. Curr Med Res Opin 1997; 14:1–20Crossref, Medline, Google Scholar
5 : Use of genome-wide association studies for drug repositioning. Nat Biotechnol 2012; 30:317–320Crossref, Medline, Google Scholar
6 : Rational drug repositioning by medical genetics. Nat Biotechnol 2013; 31:1080–1082Crossref, Medline, Google Scholar
7 : Translating genome-wide association findings into new therapeutics for psychiatry. Nat Neurosci 2016; 19:1392–1396Crossref, Medline, Google Scholar
8 : The support of human genetic evidence for approved drug indications. Nat Genet 2015; 47:856–860Crossref, Medline, Google Scholar
9 : Heritability estimates for psychotic disorders: the Maudsley twin psychosis series. Arch Gen Psychiatry 1999; 56:162–168Crossref, Medline, Google Scholar
10 : Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511:421–427Crossref, Medline, Google Scholar
11 : DGIdb: mining the druggable genome. Nat Methods 2013; 10:1209–1210Crossref, Medline, Google Scholar
12 : Polygenic overlap between schizophrenia risk and antipsychotic response: a genomic medicine approach. Lancet Psychiatry 2016; 3:350–357Crossref, Medline, Google Scholar
13 : Interactome networks and human disease. Cell 2011; 144:986–998Crossref, Medline, Google Scholar
14 : Disease networks: uncovering disease-disease relationships through the incomplete interactome. Science 2015; 347:1257601Crossref, Medline, Google Scholar
15 : Network medicine: a network-based approach to human disease. Nat Rev Genet 2011; 12:56–68Crossref, Medline, Google Scholar
16 : High-resolution network biology: connecting sequence with function. Nat Rev Genet 2013; 14:865–879Crossref, Medline, Google Scholar
17 : Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat Rev Genet 2015; 16:441–458Crossref, Medline, Google Scholar
18 : Drug-target network. Nat Biotechnol 2007; 25:1119–1126Crossref, Medline, Google Scholar
19 : DGIdb 2.0: mining clinically relevant drug-gene interactions. Nucleic Acids Res 2016; 44(D1):D1036–D1044Crossref, Medline, Google Scholar
20 : A comprehensive map of molecular drug targets. Nat Rev Drug Discov 2017; 16:19–34Crossref, Medline, Google Scholar
21 : Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 2014; 506:376–381Crossref, Medline, Google Scholar
22 : GWAS and drug targets. BMC Genomics 2014; 15(Suppl 4):S5Crossref, Medline, Google Scholar
23 : STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res 2005; 33:D433–D437Crossref, Medline, Google Scholar
24 : Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol 2011; 696:291–303Crossref, Medline, Google Scholar
25 : Schizophrenia risk from complex variation of complement component 4. Nature 2016; 530:177–183Crossref, Medline, Google Scholar
26 : Network-based in silico drug efficacy screening. Nat Commun 2016; 7:10331Crossref, Medline, Google Scholar
27 : Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science 2014; 343:506–511Crossref, Medline, Google Scholar
28 : Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet 2010; 6:1–19Crossref, Google Scholar
29 : Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet 2016; 48:624–633Crossref, Medline, Google Scholar
30 : Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 2013; 45:1452–1458Crossref, Medline, Google Scholar
31 : Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009; 41:703–707Crossref, Medline, Google Scholar
32 : ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 2009; 37:W305–W311Crossref, Medline, Google Scholar
33 : Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci 2015; 18:199–209Crossref, Medline, Google Scholar
34 : Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Am J Hum Genet 2006; 78:1011–1025Crossref, Medline, Google Scholar
35 : Interactome networks, in Handbook of Systems Biology: Concepts and Insights, 1st ed. Edited by Walhout M, Vidal M, Dekker J. New York, Elsevier, 2013, pp 45–63Google Scholar
36 : Neuropsychological decline in schizophrenia from the premorbid to the postonset period: evidence from a population-representative longitudinal study. Am J Psychiatry 2014; 171:91–101Link, Google Scholar
37 : The NIMH-MATRICS project for developing cognition-enhancing agents for schizophrenia. Dialogues Clin Neurosci 2006; 8:109–113Medline, Google Scholar
38 : Association between the polymorphic GRM3 gene and negative symptom improvement during olanzapine treatment. Schizophr Res 2005; 77:253–260Crossref, Medline, Google Scholar
39 : Molecular study of weight gain related to atypical antipsychotics: clinical implications of the CYP2D6 genotype. Rom J Morphol Embryol 2014; 55:877–884Medline, Google Scholar
40 : CYP2D6 polymorphisms and their influence on risperidone treatment. Pharm Genomics Pers Med 2016; 9:131–147Medline, Google Scholar
41 : Effect of second-generation antipsychotics on cognition: current issues and future challenges. Expert Rev Neurother 2010; 10:43–57Crossref, Medline, Google Scholar
42 : Update on new and emerging treatments for schizophrenia. Psychiatr Clin North Am 2016; 39:217–238Crossref, Medline, Google Scholar
43 : Differential contributions of prefrontal and hippocampal dopamine D(1) and D(2) receptors in human cognitive functions. J Neurosci 2008; 28:12032–12038Crossref, Medline, Google Scholar
44 : Cognition, dopamine and bioactive lipids in schizophrenia. Front Biosci (Schol Ed) 2011; 3:298–330Medline, Google Scholar
45 : Cholinergic targets for cognitive enhancement in schizophrenia: focus on cholinesterase inhibitors and muscarinic agonists. Psychopharmacology (Berl) 2004; 174:45–53Crossref, Medline, Google Scholar
46 : Role of the nicotinic acetylcholine receptor in Alzheimer’s disease pathology and treatment. Neuropharmacology 2015; 96(Pt B):255–262Crossref, Medline, Google Scholar
47 : Quantitative assessment of nicotinic acetylcholine receptor proteins in the cerebral cortex of Alzheimer patients. Brain Res Mol Brain Res 2000; 76:385–388Crossref, Medline, Google Scholar
48 : Significance of the nicotinic alpha7 receptor in cognition and antipsychotic-like behavior in the rat. Behav Brain Res 2017; 333:129–134Crossref, Medline, Google Scholar
49 : Potential role of the combination of galantamine and memantine to improve cognition in schizophrenia. Schizophr Res 2014; 157:84–89Crossref, Medline, Google Scholar
50 : Nicotinic α7 and α4β2 agonists enhance the formation and retrieval of recognition memory: potential mechanisms for cognitive performance enhancement in neurological and psychiatric disorders. Behav Brain Res 2016; 302:73–80Crossref, Medline, Google Scholar
51 : Cadherin superfamily genes: functions, genomic organization, and neurologic diversity. Genes Dev 2000; 14:1169–1180Medline, Google Scholar
52 : The protocadherin 17 gene affects cognition, personality, amygdala structure and function, synapse development and risk of major mood disorders. Mol Psychiatry (Epub ahead of print, Jan 10, 2017)Google Scholar
53 : Pathophysiology of HCN channels. Pflugers Arch 2007; 454:517–522Crossref, Medline, Google Scholar
54 : HCN1 channels: a new therapeutic target for depressive disorders? Sci Signal 2012; 5:pe44Crossref, Medline, Google Scholar
55 : HCN channels are a novel therapeutic target for cognitive dysfunction in Neurofibromatosis type 1. Mol Psychiatry 2015; 20:1311–1321Crossref, Medline, Google Scholar
56 : A proteome-scale map of the human interactome network. Cell 2014; 159:1212–1226Crossref, Medline, Google Scholar
57 : An empirical framework for binary interactome mapping. Nat Methods 2009; 6:83–90Crossref, Medline, Google Scholar
58 : Next-generation sequencing to generate interactome datasets. Nat Methods 2011; 8:478–480Crossref, Medline, Google Scholar
59 : Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol 2008; 4:682–690Crossref, Medline, Google Scholar



