Validation of Electronic Health Record Phenotyping of Bipolar Disorder Cases and Controls
Abstract
Objective:
The study was designed to validate use of electronic health records (EHRs) for diagnosing bipolar disorder and classifying control subjects.
Method:
EHR data were obtained from a health care system of more than 4.6 million patients spanning more than 20 years. Experienced clinicians reviewed charts to identify text features and coded data consistent or inconsistent with a diagnosis of bipolar disorder. Natural language processing was used to train a diagnostic algorithm with 95% specificity for classifying bipolar disorder. Filtered coded data were used to derive three additional classification rules for case subjects and one for control subjects. The positive predictive value (PPV) of EHR-based bipolar disorder and subphenotype diagnoses was calculated against diagnoses from direct semistructured interviews of 190 patients by trained clinicians blind to EHR diagnosis.
Results:
The PPV of bipolar disorder defined by natural language processing was 0.85. Coded classification based on strict filtering achieved a value of 0.79, but classifications based on less stringent criteria performed less well. No EHR-classified control subject received a diagnosis of bipolar disorder on the basis of direct interview (PPV=1.0). For most subphenotypes, values exceeded 0.80. The EHR-based classifications were used to accrue 4,500 bipolar disorder cases and 5,000 controls for genetic analyses.
Conclusions:
Semiautomated mining of EHRs can be used to ascertain bipolar disorder patients and control subjects with high specificity and predictive value compared with diagnostic interviews. EHRs provide a powerful resource for high-throughput phenotyping for genetic and clinical research.
Since 2006, genome-wide association studies (GWAS) have identified specific genetic variants underlying a range of common medical disorders. At the same time, these findings have demonstrated that a rate-limiting challenge for successful gene identification is the availability of large populations of case and control subjects. For example, the detection of loci influencing complex disorders such as schizophrenia and diabetes required tens of thousands of such individuals (1, 2). The evidence thus far suggests that the genetic architecture of psychiatric disorders involves multiple loci of modest effect (3). Emerging evidence from GWAS of bipolar disorder has been promising (4), but there is now an urgent need for the collection and genetic analyses of much larger cohorts than have been studied to date in order to identify the common and rare variants that underlie the substantial heritability of bipolar disorder.
The increasing utilization of electronic health records (EHRs) provides new opportunities for epidemiologic and genetic research. A ready repository of clinical and phenotypic data contained in health system EHRs can enable low-cost population-based studies of unprecedented size. A growing number of studies have mined these data for a range of applications, including pharmacovigilance (5–8) and genetic association studies (9–11). In addition to the use of structured codified data (e.g., diagnostic codes, demographic variables), text mining by natural language processing allows the accrual and analysis of detailed, longitudinal clinical data for research purposes (12).
Support for the validity of EHR-based diagnosis has emerged from GWAS in which previously established gene associations have been detected in independent samples by using phenotypes derived from EHRs (11, 13–15). However, the use of informatics-based phenotyping for psychiatric disorders presents special challenges. Unlike most other classes of medical illness, psychiatric disorders lack established biological markers of diagnosis. Clinical diagnosis in psychiatry relies on constellations of self-reported symptoms and behavioral observation. There is widespread concern that misclassification may occur without extensive, validated diagnostic methods. Given this, the gold standard in clinical, epidemiologic, and genetic studies of psychopathology has been direct assessment by trained observers or clinicians using structured or semistructured diagnostic interviews. However, such methods are costly and labor-intensive. Alternative methods have been validated (e.g., schizophrenia diagnosis based on diagnostic codes in a Swedish Hospital Discharge Registry [3]), but such methods have not been widely used.
In the present study, we sought to evaluate the validity of EHR-based case and control ascertainment of bipolar disorder. We defined a set of algorithms to extract diagnostic data from the EHRs of a large health care system. The algorithms included one based on natural language processing and several based on coded variables. We assessed the diagnostic validity of each algorithm against the gold standard of in-person semistructured interviews conducted by trained clinical researchers. Here we show that high levels of diagnostic specificity and positive predictive values (PPVs) for bipolar disorder case and control subjects are achievable by means of high-throughput EHR data mining.
Method
This study was conducted as part of the International Cohort Collection for Bipolar Disorder (ICCBD), an international consortium designed to collect a large sample (N=19,000 case and 19,000 control subjects) for genetic studies of bipolar disorder. The Massachusetts General Hospital site of the ICCBD aimed to collect DNA from 4,500 cases and 4,500 controls by linking discarded blood samples to de-identified EHR data.
Data Source and Population
A schematic diagram of the study is presented in Figure 1. Our primary data source was the Partners Healthcare Research Patient Data Registry, which spans more than 20 years of data from 4.6 million patients. The database contains over 227 million encounters, 193 million ICD-9 diagnoses, 105 million medications, 200 million procedures, 852 million laboratory values, and over 55 million unstructured clinical notes, which are a combination of outpatient visit notes, inpatient discharge summaries, radiology reports, and others. The registry population is approximately 55% female and 72% Caucasian and has an average age of 45.7 years (SD=23.2).
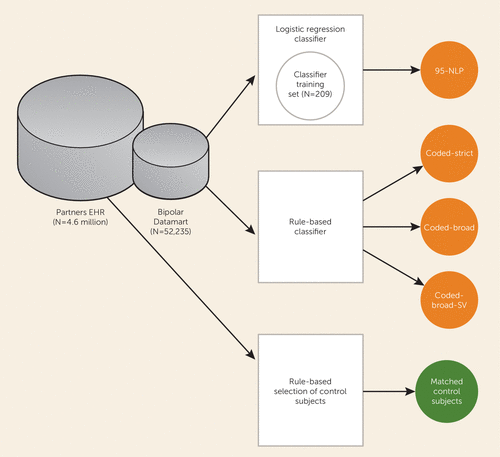
FIGURE 1. Procedure for Validating Electronic Health Record Phenotyping of Bipolar Disorder and Control Subjectsa
a Partners EHR: electronic health records in Partners Healthcare Research Patient Data Registry. Datamart: electronic billing data or outpatient medical records at Massachusetts General Hospital, Brigham and Women’s Hospital, and McLean Hospital. 95-NLP: probabilistic algorithm with 95% specificity based on natural language processing. SV: single visit.
Patients with at least one diagnosis of bipolar disorder (ICD-9 and DSM-IV-TR codes 296.4*–296.8*) or manic disorder (ICD 296.0*–296.1*) in the billing data or outpatient medical records at Massachusetts General Hospital, Brigham and Women’s Hospital, or McLean Hospital were selected for inclusion in a data set, referred to as a “datamart.” The datamart consisted of all electronic records from 52,235 patients analyzed with the Informatics for Integrating Biology and the Bedside (i2b2) Workbench software (i2b2 v1.6.04; https://www.i2b2.org/software/index.html#) (16). The i2b2 system is a scalable computational framework for managing health data, and Workbench facilitates data analysis and visualization (17). Billing code data were available for all public and private payers. Medication data were available from both medications dispensed by an inpatient pharmacy (27%) and medications prescribed in the EHR (73%). The Partners HealthCare System institutional review board approved all aspects of this study.
Clinician Chart Review to Establish Gold Standard
From the bipolar datamart, a random sample of 209 patients with at least one outpatient psychiatric diagnostic evaluation note, inpatient or emergency psychiatric consultation note, or discharge summary from a psychiatric inpatient unit were selected for chart review. Three experienced, board-certified psychiatrists (J.W.S., R.H.P., M.N.V.) reviewed all psychiatric notes in the patient’s record and arrived at a consensus diagnostic status of bipolar disorder, not bipolar disorder, or not enough information. Review guidelines for assigning diagnostic status were adapted from the DSM-IV criteria for bipolar disorder. A confidence level of high, moderate, or low was also assigned to each classification to denote the level of evidence supporting the diagnosis (see Figure S1 in the data supplement accompanying the online version of this article).
Classification Algorithm Using Natural Language Processing
During the chart review, clinicians also identified terms in the narratives that were either consistent or inconsistent with a diagnosis of bipolar disorder (e.g., “increasing racing thoughts” is consistent with bipolar disorder, and “no history of mania” is inconsistent). The instances of related diagnoses, encounters, procedures, and medications from the structured medical record were also identified as consistent or inconsistent with bipolar disorder (the full list of features is available in the Data S1 section of the online data supplement). These terms were subsequently extracted from each narrative note with natural language processing using the HITEx platform (18), which identifies terms using regular expressions (flexible matching) and applies negation and context algorithms to filter inappropriate matches. The presence or absence of a term then becomes a feature of each note, which can be used in classification algorithms.
We used the clinician-reviewed classifications to train models to predict the probability of a bipolar diagnosis or no bipolar diagnosis with a confidence level of moderate or high at each visit on the basis of a logistic regression classifier with the adaptive least absolute shrinkage and selection operator (LASSO) procedure. The adaptive LASSO procedure simultaneously identifies important features and provides stable estimates of the model parameters (19). It is often applied in high-dimensional data sets to select the more useful subset of features for modeling because it shrinks the coefficients of noninformative features (covariates) to zero. The optimal penalty parameter was determined on the basis of the Bayesian information criterion. We first trained a note-level model to predict the probability of bipolar disorder given feature information from each note. Since the amount of diagnostic information contained in an evaluation note could differ substantially from that in a follow-up note, we trained a second logistic regression model using the note-level predicted bipolar disorder probability and the type of clinical note as features. This second model aggregates longitudinal information to classify bipolar disorder at the patient level.
Rule-Based Classification Algorithms for Bipolar Disorder
Because the regression classification algorithm required that patients have electronic psychiatric clinical notes, which were widely adopted only in 2007, we developed additional rule-based classifiers that rely solely on coded diagnostic, encounter, and medication information, which have been recorded uniformly since 1998. Three coded rule-based algorithms—coded-broad, coded-strict, coded-strict-single-visit (coded-strict-SV)—for identifying patients with bipolar disorder were developed on the basis of the patient’s diagnostic and treatment history. Table 1 outlines the criteria for each rule-based algorithm.
| Ascertainment Method | Classifier Description | ICD–9 Diagnosis Criteria | Medication Criteria | Visit Criteria | Other Criteria |
|---|---|---|---|---|---|
| 95-NLP | Probabilistic algorithm with 95% specificity based on coded feature and natural language processing (NLP) feature | ≥2 BD diagnoses AND no diagnosis of MDD, SCZ, SZA, or OAS, unless two most recent diagnoses are BD | None | ≥1 psychiatric visit with electronic clinical note in EHR | Selected by logistic regression model |
| Coded-strictb | ≥3 diagnoses and BD-specific treatment | ≥3 BD diagnoses ANDeither 1) no diagnosis of MDD, SCZ, SZA, or OAS, unless two most recent diagnoses are BD, or 2) number of MDD, SCZ, SZA, or OAS diagnoses is greater (>50%) than number of BD diagnoses | Li or VPA within 1 year of a BD diagnosis | ≥2 visits at BD specialty clinic (with BD diagnosis) | |
| Coded-broad | ≥2 diagnoses and treatment | ≥2 BD diagnoses (at separate visits >1 month apart) ANDeither 1) no diagnosis of MDD, SCZ, SZA, or OAS, unless two most recent diagnoses are BD, or 2) number of MDD, SCZ, SZA, or OAS diagnoses is greater (>50%) than number of BD diagnoses | ≥2 BD medications—Li, VPA, or other mood stabilizer (risperidone, olanzapine, quetiapine, ziprasidone, aripiprazole, or carbamazepine)—within 1 year of a BD diagnosis | None | |
| Coded-broad-SV | ≥2 diagnoses and treatment in single episode of inpatient or outpatient care | ≥2 BD diagnoses during single inpatient or outpatient episode ANDeither 1) no diagnosis of MDD, SCZ, SZA, or OAS, unless two most recent diagnoses are BD, or 2) number of MDD, SCZ, SZA, or OAS diagnoses is greater (>50%) than number of BD diagnoses | ≥2 BD drugs—Li, VPA, or other mood stabilizer (risperidone, olanzapine, quetiapine, ziprasidone, aripiprazole, or carbamazepine)—within 1 year of a BD diagnosis | None | |
| Control subjectsb | Matched to case subjects on age, race, and gender | Age >30 years AND no history of any mental health disorder diagnosis | No history of any mental health disorder medications | ≥2 encounters at Mass Gen, Brigham/Women’s, or McLean |
TABLE 1. Probabilistic and Rule-Based Algorithms for Classifying Bipolar Disordera
Rule-Based Classification Algorithm for Control Subjects
We identified a cohort of control patients who were at least 30 years old and had no ICD-9 codes or history of medications related to a psychiatric or neurological condition. We selected 1.2 million patients meeting these criteria in the research patient data registry for a control pool. The control patients were then matched 15:1 to the algorithm-classified case patients on the basis of age, gender, race/ethnicity, and health care utilization (number of facts) by using a standard frequency matching approach.
Validation Clinical Study
Bipolar disorder case and control patients identified by the algorithms underwent semistructured diagnostic interviews using the Structured Clinical Interview for DSM-IV (SCID-IV) by an experienced doctoral-level clinician blinded to the classifier diagnosis and method of selecting the cohort. Interviewers were required to undergo formal SCID training (as recommended at www.SCID4.org), which was documented for each interviewer. This included careful review of the SCID User’s Guide, instructions, and interview; viewing seven SCID training DVDs; and documenting concordant diagnoses with two SCID training interviews.
Individuals selected by the classification algorithms were invited by mail to participate in the in-person validation study. Subjects were ascertained by a hierarchical application of the algorithms such that they were selected on the basis of the most stringent algorithm for which they met the case definition (95-NLP > coded-strict > coded-broad > coded-broad-SV). The SCID assessment was completed by 190 patients, including 45 patients selected by the 95-NLP probabilistic algorithm; 59 selected by the coded-strict, 31 by the coded-broad, and eight by the coded-broad-SV algorithms; and 20 matched control subjects. To further preserve clinician blinding, we also recruited 27 individuals from advertisements in community clinics at Massachusetts General Hospital who reported a previous diagnosis of schizophrenia or major depression, two disorders commonly considered in the differential diagnosis of bipolar disorder.
Extraction of Subphenotypes
For cases, we aimed to classify relevant subphenotypes associated with bipolar disorder: age at bipolar disorder onset, bipolar disorder subtype, family history of bipolar disorder, and history of: alcohol dependence, drug dependence, suicide attempt, psychosis, or panic disorder/agoraphobia. Two board-certified psychiatrists (J.W.S., R.H.P.) manually reviewed 620 notes to identify important terms (features) indicative of each subphenotype. Each feature was extracted from the notes by using the HITEx system (18). The gold standard subphenotype classification was based on results of the SCID direct interview and was used to train algorithms using the extracted features. All case patients were used in the training phase regardless of whether they received a SCID diagnosis of bipolar disorder. We trained a separate model for each subphenotype by using the LASSO regression procedure with 10-fold cross-validation. There were two exceptions to the above procedure. Age at onset was categorized into early onset (age <18), typical onset (age 18–40), and late onset (age >40); bipolar subtype was categorized into bipolar disorder I, bipolar disorder II, other bipolar disorder, and schizoaffective disorder, bipolar type. To validate the categorization of these two subphenotypes, the research coordinator reviewed text from 701 notes that included explicit mention of bipolar disorder subtype or age at onset and assigned the appropriate category.
Statistical Analysis for Validation Study
For the algorithm using natural language processing, performance of the logistic regression model was assessed by using receiver operating curve (ROC) analysis for models in which specificity was set at the desired threshold of 95%. The overall performance of this algorithm, referred to as 95-NLP, was summarized by using the area under the ROC curve (AUC). Performance of the case and control classification compared with the in-person validation study was assessed by using the PPV for the algorithm classification relative to the SCID classification. The PPV for cases was calculated as the proportion of cases diagnosed as bipolar (bipolar I, bipolar II, other bipolar, or schizoaffective disorder, bipolar type) by SCID interview given an algorithm diagnosis of bipolar disorder. This PPV is based on a base population defined by inclusion in the bipolar datamart (i.e., having at least one billing code for bipolar disorder or manic disorder). Because cases selected by one algorithm (e.g., 95-NLP) might also be classifiable by another algorithm (e.g., coded-strict), we also calculated the PPV by allowing each case to be included for any algorithm capable of classifying the case. For example, if a subject was ascertained with the 95-NLP algorithm but also met the criteria for bipolar disorder according to the coded-strict and coded-broad rules, she would be included in calculations of PPV for all three definitions. This “nonhierarchical” PPV provides an estimate of the diagnostic performance of each algorithm regardless of the algorithm by which subjects were ascertained. The PPV for control subjects represents the proportion of individuals classified as control subjects (no bipolar disorder diagnosis) by SCID interview given an algorithm classification as a control. For subphenotype assessment, PPVs were calculated against the SCID interview gold standard.
Results
After manual review of 612 notes from the 209 randomly selected patients in the bipolar datamart, 132 patients were classified as “bipolar” (37% with high confidence, 26% with moderate confidence, 37% with low confidence), 69 were classified as “not bipolar” (36% with high, 35% with moderate, and 29% with low confidence), and eight were classified as “insufficient information.” We identified 401 terms relevant to bipolar disorder to be used as features in the model training. An additional 13 relevant coded terms from the EHR, such as those relating to sex and past prescription of lithium, were also included as features.
Of the 414 features identified for model training, the adaptive LASSO selected 13 features for bipolar disorder classification (Table 2). The final model for classifying each note as indicating a bipolar disorder diagnosis yielded an AUC of 0.93 (SE=0.01), with a sensitivity of 0.53 when the specificity was set at 0.95 (Figure 2). The AUC for classifying an individual as having bipolar disorder or not across notes and other longitudinal data was 0.82 (SE=0.03). After running the logistic regression classifier on datamart patients with sufficient clinical narratives, an initial set of 1,776 patients were selected as having bipolar disorder. Patients in the datamart not classified by the probabilistic algorithm were eligible for classification by the rule-based algorithms. In this process, 11,492 patients were selected by the coded-strict algorithm, 3,381 by the coded-broad algorithm, and 5,220 by the coded-broad-SV algorithm, and 296,356 control subjects with no psychiatric or neurologic disorders were matched to the case subjects (Table 3).
| Model Term | 95-NLP Model Beta Weight | Variable Type | Description |
|---|---|---|---|
| Intercept | –1.1360 | Linear model intercept | |
| Bipolar ICD–9 code at visit | 1.4571 | Coded | Presence of a bipolar ICD-9 code (296.0-1 or 296.4-8) at visit |
| Psychopharmacology note | 0.8018 | NLP | Note describes a psychopharmacology visit |
| Mood stabilizer at visit | 0.4400 | Coded | Patient was prescribed a mood stabilizer at the visitb |
| hx.bipolar.disorder | 0.4049 | NLP | Mention of a history of bipolar disorder in text of note |
| dx.bipolar.disorder | 0.2567 | NLP | Mention of a diagnosis of bipolar disorder in text of note |
| bi.med...anticonvulsant..non.vpa | –0.1262 | NLP | Mention of an anticonvulsant (other than valproic acid) in text of note |
| anxiety.disorders | –0.1466 | NLP | Mention of anxiety disorder in text of note |
| dx.psychotic.disorder | –0.1966 | NLP | Mention of a psychotic disorder in text of note |
| neuro.cognitive.impairment | –0.2201 | NLP | Mention of neurocognitive impairment in text of note |
| X90801_visit | –0.6617 | Coded | Psychological Diagnostic Interview Examination at visit (CPT-4 90801) |
| dx.schizoaffective | –0.7460 | NLP | Mention of schizoaffective in text of note |
| dx.depression | –2.3788 | NLP | Mention of depression in text of note |
TABLE 2. Patient or Visit Features Identified for Training a Natural Language Processing Model (95-NLP) to Classify Bipolar Disordera
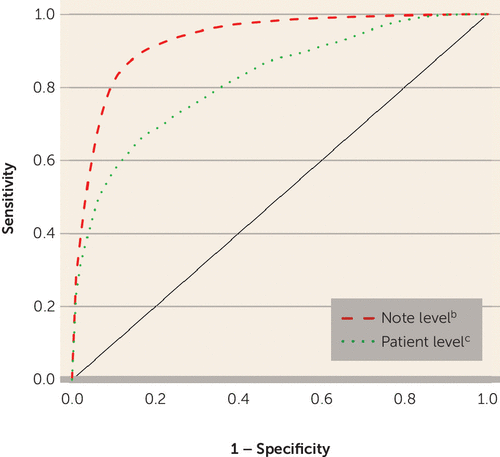
FIGURE 2. Area Under the Curve (AUC) for a Natural Language Processing Model (95-NLP) of Classifying Bipolar Disordera
a 95-NLP: probabilistic algorithm with 95% specificity based on natural language processing. Receiver operator curves are presented for the classification of bipolar disorder based on feature information from each clinical note (note level) and for the classification of bipolar disorder across notes for a given patient (patient level).
b Relaxed adaptive least absolute shrinkage and selection operator (LASSO): AUC=0.931.
c Logistic regression: AUC=0.823.
| Age (years) | Race/Ethnicity (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Algorithm and Groupb | N | Mean | SD | White | Black | Hispanic | Asian | Other/Unknown | Female (%) |
| 95-NLP | |||||||||
| Classified | 1,776 | 40.8 | 15.6 | 77 | 8 | 6 | 2 | 6 | 55 |
| Validation subset | 45 | 42.8 | 11.9 | 80 | 9 | 4 | 4 | 2 | 60 |
| Coded-strict | |||||||||
| Classified | 11,492 | 48.5 | 16.4 | 78 | 5 | 4 | 1 | 12 | 55 |
| Validation subset | 59 | 54.2 | 13.3 | 83 | 12 | 2 | 2 | 2 | 56 |
| Coded-broad | |||||||||
| Classified | 3,381 | 42.8 | 16.9 | 77 | 7 | 6 | 1 | 8 | 63 |
| Validation subset | 31 | 45.5 | 16.2 | 84 | 6 | 10 | 0 | 0 | 61 |
| Coded-broad-SV | |||||||||
| Classified | 5,220 | 45.4 | 17.1 | 75 | 5 | 4 | 1 | 14 | 57 |
| Validation subset | 8 | 51.9 | 9.1 | 100 | 0 | 0 | 0 | 0 | 75 |
| Control subjects | |||||||||
| Classified | 296,356 | 48.5 | 12.7 | 78 | 8 | 7 | 1 | 5 | 56 |
| Validation subset | 20 | 50.9 | 12.4 | 90 | 5 | 0 | 5 | 0 | 55 |
TABLE 3. Demographic Characteristics of Patients Classified by Probabilistic and Rule-Based Algorithms as Having Bipolar Disorder in the Datamart Samplea
According to the SCID gold standard, the cases selected by 95-NLP yielded a PPV of 0.85 (95% confidence interval [CI]: 0.72–0.93) (Table 4). The coded algorithms resulted in PPVs of 0.79 (95% CI: 0.67–0.87) for cases selected by the coded-strict algorithm, 0.62 (95% CI: 0.43–0.78) for coded-broad, and 0.50 (95% CI: 0.22–0.78) for coded-broad-SV. No patients selected by the control rules were given a SCID diagnosis of bipolar disorder (PPV: 1.00, 95% CI: 0.84–1.00). As shown at the bottom of Table 4, when results were calculated on the basis of nonhierarchical rules (that is, classifying subjects according to all rules for which they met criteria), the PPVs for the coded-strict and coded-broad algorithms increased substantially.
| SCID-I Diagnosis | Algorithm Validity | |||||||
|---|---|---|---|---|---|---|---|---|
| Algorithma | N | Bipolar Disorder I | Bipolar Disorder II | Bipolar Disorder, Other | Schizoaffective Disorder, Bipolar Type | No Bipolar Disorder | Positive Predictive Value | 95% CI |
| Subjects classified by hierarchical algorithm | ||||||||
| Case subjects | ||||||||
| 95-NLP | 47 | 31 | 3 | 5 | 1 | 7 | 0.85 | 0.72–0.93 |
| Coded-strict | 62 | 37 | 9 | 2 | 1 | 13 | 0.79 | 0.67–0.87 |
| Coded-broad | 26 | 12 | 1 | 3 | 0 | 10 | 0.62 | 0.43–0.78 |
| Coded-broad-SV | 8 | 3 | 1 | 0 | 0 | 4 | 0.50 | 0.22–0.78 |
| Matched control subjects | 20 | 0 | 0 | 0 | 0 | 20 | 1.00 | 0.84–1.00 |
| Case subjects classified by nonhierarchical rulesb | ||||||||
| 95-NLP | 66 | 46 | 4 | 6 | 1 | 9 | 0.86 | 0.72–0.93 |
| Coded-strict | 98 | 63 | 10 | 8 | 1 | 16 | 0.84 | 0.75–0.90 |
| Coded-broad | 129 | 78 | 12 | 11 | 2 | 26 | 0.80 | 0.72–0.86 |
| Coded-broad-SV | 8 | 3 | 1 | 0 | 0 | 4 | 0.50 | 0.22–0.78 |
TABLE 4. Validation of Probabilistic and Rule-Based Algorithms for Classifying Bipolar Disorder, Relative to Structured Diagnostic Interviews
Table 5 provides positive and negative predictive values for each of the eight relevant subphenotype algorithms. Applying these algorithms to the selected bipolar disorder cases, we identified a history of alcohol abuse in 54% of the patients, a history of substance abuse in 40%, a history of psychosis in 35%, a past suicide attempt in 10%, and a history of panic disorder in 42%. In addition, 75% of the case subjects were identified as having bipolar I disorder, and 35% of the case subjects had an identified family history of bipolar disorder. Where the age at onset was known, 46% of patients were identified as having an early onset (age <18) and 10% as having a late onset (age >40).
| Algorithm Validity | |||||||
|---|---|---|---|---|---|---|---|
| Subphenotype | N | Categories | Overall Area Under the Curve | Positive Predictive Value | 95% CI | Negative Predictive Value | 95% CI |
| Alcohol abuse | 143 | Present, absent | 0.810 | 0.89 | 0.83–0.93 | 0.53 | 0.42–0.65 |
| Substance abuse | 143 | Present, absent | 0.647 | 0.81 | 0.69–0.89 | 0.67 | 0.59–0.74 |
| Psychosis | 140 | Present, absent | 0.674 | 0.72 | 0.59–0.83 | 0.70 | 0.60–0.78 |
| Panic/agoraphobia | 143 | Present, absent | 0.731 | 0.83 | 0.72–0.91 | 0.67 | 0.56–0.77 |
| Suicide attempt | 139 | Present, absent | 0.825 | 0.92 | 0.67–0.99 | 0.92 | 0.86–0.96 |
| Family history of bipolar disorder | 105 | Present, absent | 0.695 | 0.73 | 0.57–0.84 | 0.70 | 0.57–0.84 |
| Bipolar subtypea | 100 | Bipolar disorder I; bipolar disorder II; bipolar disorder not otherwise specified; schizoaffective disorder, bipolar type | 0.77 | 0.68–0.84 | |||
| Age at onset of bipolar disordera | 100 | Early (<18), typical (18–40), late (>40), onset unknown | 0.94 | 0.88–0.97 | |||
TABLE 5. Validation of an Algorithm for Classifying Bipolar Disorder Subphenotypes, Relative to Structured Diagnostic Interviews
Discussion
Clinical characterization in psychiatric research has traditionally been an expensive and labor-intensive proposition involving lengthy diagnostic interviews. The expanding availability of EHRs offers a new and powerful alternative for the collection of diagnostic and outcome data. In the realm of genetic research, the accrual of large samples of case and control subjects has become a rate-limiting challenge for the discovery of risk variants. Prior studies by our group and others have supported the validity of EHR-based phenotyping by replicating genetic and epidemiologic findings by means of these methods (10, 11, 15). We have also previously demonstrated the utility of longitudinal EHR phenotyping for pharmacovigilance, neuroimaging, and treatment outcome research (5, 6, 20–22). However, the present study provides direct validation of informatic-based ascertainment by comparing diagnoses derived from EHRs to a gold standard of traditional clinician-based interviews.
Several findings of this study warrant highlighting. First, we found that text mining of medical records using natural language processing can be used to develop highly specific and predictive diagnostic algorithms that are comparable to those achieved by direct interview. In the model-training phase, we derived an algorithm using natural language processing that had 95% specificity and high predictive validity (AUC=0.82) compared with expert clinician-derived diagnoses of bipolar disorder by manual chart review. In the direct-interview validation phase, our natural language processing algorithm demonstrated high predictive validity compared with blinded semistructured clinical interviews (PPV=0.85). This degree of diagnostic accuracy is particularly notable in the context of the interrater reliability of standard diagnostic interviews themselves. For example, the DSM-5 field trials had a pooled kappa of only 0.56 for bipolar I disorder when patients were evaluated by two independent clinicians within 2 weeks of each other (23) (studies using earlier diagnostic criteria achieved higher though still imperfect reliability estimates [24, 25]). Thus, some degree of diagnostic imprecision is expected and likely unavoidable.
We also obtained excellent PPVs for certain algorithms based on coded EHR data. The coded-strict algorithm, which required a history of multiple bipolar disorder diagnoses and either treatment at a bipolar disorder specialty clinic or prescription of lithium or valproate, achieved a PPV of 0.79 (rising to 0.84 when nonhierarchical rules were used). In addition, our diagnostic rule for ascertainment of control subjects, comprising multiple filters to exclude psychopathology, yielded a PPV of 1.0.
Less robust performance was seen for the remaining diagnostic rules, which relied on a broader set of criteria. The coded-broad definition required at least two bipolar disorder diagnoses, a predominance of bipolar disorder diagnoses over diagnoses of other psychotic disorders or depression, and treatment with lithium, valproate, or antipsychotic medication. The PPV for this definition was 0.62 but rose to 0.80 when the nonhierarchical classification was used. The coded-broad-SV definition was identical except that the coded bipolar disorder diagnoses could have been given less than 1 month apart. It is noteworthy that these criteria are still more stringent than those often used in population-based studies that rely on claims data in which one or two instances of a diagnostic code are used to define cases. Indeed, our results suggest that studies relying on such claims-based criteria are likely to include a substantial proportion of false positives. The prospective, longitudinal nature of EHRs also provides a critical advantage for diagnosis. For example, longitudinal studies indicate that as many as 15% of bipolar cases are later diagnosed as schizophrenia or schizoaffective disorder (26, 27) and nearly 40% of individuals with psychotic depression later receive a non-mood-disorder diagnosis (28). Thus, claims-based studies that rely on the presence of a single diagnostic code may result in substantial misclassification.
We also examined the reliability of several subphenotypes and comorbidities that are relevant for genetic subtyping. The PPV statistics comparing informatic-based diagnosis to diagnostic interview demonstrate that such finer-grained phenotyping by EHR-based algorithms is a viable approach. However, ambiguous information for some of these phenotypes (e.g., a lack of an affirmative statement or negation in the record) meant that we were unable to classify a portion of cases with respect to these subphenotypes.
Our high-throughput informatics-based phenotyping approach was designed to allow the rapid accrual of diagnostic data and blood samples for genetic analysis. We used these definitions to ascertain case and control subjects for the ICCBD consortium by linking phenotypic data to discarded blood samples as previously described (11). In brief, case and control medical record numbers are submitted to the Partners HealthCare Crimson system, which allows prospective collection of discarded samples. Acting as an “honest broker,” Crimson matches deidentified phenotypic data to discarded blood samples. Using the case/control definitions described in this study, we collected approximately 4,500 subjects with bipolar disorder and 5,000 control subjects over 3 years. The control blood samples were collected in 10 weeks. Prior simulations have demonstrated that EHR-based ascertainment and sample collection for genetic studies using the i2b2 system provide an approximate 10-fold reduction in cost compared with standard methods (29). In sum, the framework we have validated here provides a high-throughput and cost-effective engine for genetic discovery that is exportable to other health care systems (30).
There are several limitations of our study. First, the precision of our PPV estimates is limited by the sample size. In particular, we had difficulty recruiting subjects who fell into the coded-broad-SV category, and the 95% CI around our point estimate for PPV is correspondingly broad. Recruitment of these subjects was undoubtedly more difficult because of the nature of the phenotype definition. Specifically, while these participants received more than one bipolar disorder diagnosis, the diagnoses occurred during a single episode of inpatient or outpatient care. This likely captured individuals who are no longer in the health care system and were thus more likely to be lost to follow-up. Second, the applicability of our methods to other health care systems may vary depending on informatics infrastructure. Fortunately, EHR mining is increasingly widespread, including through the growing network of systems that have adopted the i2b2 platform (12, 31). Second, we included cases of bipolar disorder not otherwise specified in our definition of bipolar disorder cases, although some genetic studies have excluded such cases. However, such cases have been included in numerous recent large-scale bipolar disorder GWAS (e.g., those described in references 4 and 32). Classifying these cases as indeterminate has a negligible effect on the PPVs shown in Table 4, reducing them by 0%−3%.
In sum, our results support the validity and utility of informatic-based phenotyping for psychiatric research. It is important that the EHR ascertainment of bipolar disorder case and control subjects was highly concordant with the gold standard of in-person diagnostic interviews. The best-performing case definition algorithm made use of natural language processing, but we demonstrated that, when guided by clinical expertise, algorithms that extract coded EHR data can also yield valid phenotypes. In addition to being used on their own, EHR algorithms could be useful as a preliminary screening step to ascertain an “enriched” set of case or control subjects followed by more traditional direct interview phenotyping. With the increasingly widespread implementation of EHRs, this study supports the application of high-throughput in silico phenotyping for epidemiologic, genetic, and clinical research.
1 , et al: Genome-wide association study identifies five new schizophrenia loci. Nat Genet 2011; 43:969–976Crossref, Medline, Google Scholar
2 , et al: Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008; 40:638–645Crossref, Medline, Google Scholar
3 , et al: Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet 2013; 45:1150–1159Crossref, Medline, Google Scholar
4 : Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 2011; 43:977–983Crossref, Medline, Google Scholar
5 , et al: Antidepressant response in patients with major depression exposed to NSAIDs: a pharmacovigilance study. Am J Psychiatry 2012; 169:1065–1072Link, Google Scholar
6 , et al: Incident user cohort study of risk for gastrointestinal bleed and stroke in individuals with major depressive disorder treated with antidepressants. BMJ Open 2012; 2:e000544Crossref, Medline, Google Scholar
7 , et al: Detecting drug interactions from adverse-event reports: interaction between paroxetine and pravastatin increases blood glucose levels. Clin Pharmacol Ther 2011; 90:133–142Crossref, Medline, Google Scholar
8 , et al: Rapid identification of myocardial infarction risk associated with diabetes medications using electronic medical records. Diabetes Care 2010; 33:526–531Crossref, Medline, Google Scholar
9 , et al: Predicting clopidogrel response using DNA samples linked to an electronic health record. Clin Pharmacol Ther 2012; 91:257–263Crossref, Medline, Google Scholar
10 , et al: Associations of autoantibodies, autoimmune risk alleles, and clinical diagnoses from the electronic medical records in rheumatoid arthritis cases and non-rheumatoid arthritis controls. Arthritis Rheum 2013; 65:571–581Crossref, Medline, Google Scholar
11 , et al: Genetic basis of autoantibody positive and negative rheumatoid arthritis risk in a multi-ethnic cohort derived from electronic health records. Am J Hum Genet 2011; 88:57–69Crossref, Medline, Google Scholar
12 : Using electronic health records to drive discovery in disease genomics. Nat Rev Genet 2011; 12:417–428Crossref, Medline, Google Scholar
13 , et al: Facilitating pharmacogenetic studies using electronic health records and natural-language processing: a case study of warfarin. J Am Med Inform Assoc 2011; 18:387–391Crossref, Medline, Google Scholar
14 , et al: Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet 2010; 86:560–572Crossref, Medline, Google Scholar
15 , et al: A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum Genet 2014; 133:95–109Crossref, Medline, Google Scholar
16 , et al: Architecture of the open-source clinical research chart from informatics for integrating biology and the bedside, in American Medical Informatics Association Annual Symposium 2007: Biomedical and Health Informatics: From Foundations to Applications to Policy. Edited by Teich JM. Red Hook, NY, Curran, 2010, p 548Google Scholar
17 : A translational engine at the national scale: informatics for integrating biology and the bedside. J Am Med Inform Assoc 2012; 19:181–185Crossref, Medline, Google Scholar
18 , et al: Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak 2006; 6:30Crossref, Medline, Google Scholar
19 : On the Adaptive Elastic-Net with a Diverging Number of Parameters. Ann Stat 2009; 37:1733–1751Crossref, Medline, Google Scholar
20 , et al: Using electronic medical records to enable large-scale studies in psychiatry: treatment resistant depression as a model. Psychol Med 2012; 42:41–50Crossref, Medline, Google Scholar
21 , et al. Limbic system white matter microstructure and long-term treatment outcome in major depressive disorder: a diffusion tensor imaging study using legacy data. World J Biol Psychiatry 2014; 15:122–134Crossref, Medline, Google Scholar
22 , et al: Feasibility of studying brain morphology in major depressive disorder with structural magnetic resonance imaging and clinical data from the electronic medical record: a pilot study. Psychiatry Res 2013; 211:202–213Crossref, Medline, Google Scholar
23 , et al: DSM-5 field trials in the United States and Canada, part II: test-retest reliability of selected categorical diagnoses. Am J Psychiatry 2013; 170:59–70Link, Google Scholar
24 , et al: Diagnostic reliability of bipolar II disorder. Arch Gen Psychiatry 2002; 59:736–740Crossref, Medline, Google Scholar
25 , et al: Diagnostic accuracy and confusability analyses: an application to the Diagnostic Interview for Genetic Studies. Psychol Med 1996; 26:401–410Crossref, Medline, Google Scholar
26 , et al: Diagnostic shifts during the decade following first admission for psychosis. Am J Psychiatry 2011; 168:1186–1194Link, Google Scholar
27 : Bipolar disorder, schizoaffective disorder, and schizophrenia overlap: a new comorbidity index. J Clin Psychiatry 2009; 70:1432–1438Crossref, Medline, Google Scholar
28 , et al: Diagnostic consistency of major depression with psychosis across 10 years. J Clin Psychiatry 2011; 72:1207–1213Crossref, Medline, Google Scholar
29 , et al: Instrumenting the health care enterprise for discovery research in the genomic era. Genome Res 2009; 19:1675–1681Crossref, Medline, Google Scholar
30 , et al: Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J Am Med Inform Assoc 2012; 19(e1):e162–e169Crossref, Medline, Google Scholar
31 , et al: Technical desiderata for the integration of genomic data into electronic health records. J Biomed Inform 2012; 45:419–422Crossref, Medline, Google Scholar
32 , et al: Meta-analysis of genome-wide association data of bipolar disorder and major depressive disorder. Mol Psychiatry 2011; 16:2–4Crossref, Medline, Google Scholar



