Identification and Replication of a Combined Epigenetic and Genetic Biomarker Predicting Suicide and Suicidal Behaviors
Abstract
Objective:
Reliable identification of individuals at high risk for suicide is a priority for suicide prevention. This study was conducted to identify genes exhibiting epigenetic variation associated with suicide and suicidal behaviors.
Method:
Genome-wide DNA methylation profiling was employed separately on neuronal and glial nuclei in a discovery set of postmortem brains from the National Institute of Child Health and Human Development to identify associations with suicide. Pyrosequencing-based validation was conducted in prefrontal cortical tissue in cohorts from the Stanley Medical Research Institute and Harvard Brain Bank at McLean Hospital and peripheral blood from three living groups. Functional associations with gene expression, stress and anxiety, and salivary cortisol were assessed.
Results:
The DNA methylation scan identified an additive epigenetic and genetic association with suicide at rs7208505 within the 3′ untranslated region of the SKA2 gene independently in the three brain cohorts. This finding was replicated with suicidal ideation in blood from three live cohorts. SKA2 gene expression was significantly lower in suicide decedents and was associated with genetic and epigenetic variation of rs7208505, possibly mediated by interaction with an intronic microRNA, miR-301a. Analysis of salivary cortisol measurements suggested that SKA2 epigenetic and genetic variation may modulate cortisol suppression, consistent with its implicated role in glucocorticoid receptor transactivation. SKA2 significantly interacted with anxiety and stress to explain about 80% of suicidal behavior and progression from suicidal ideation to suicide attempt.
Conclusions:
These findings implicate SKA2 as a novel genetic and epigenetic target involved in the etiology of suicide and suicidal behaviors.
Suicide is a complex, heterogeneous phenotype as well as an intractable public health problem, with the overall annual suicide rate remaining stable over the past 60 years at around 10 to 12 per 100,000 (1). The National Action Alliance for Suicide Prevention has set out to build a research agenda with the potential to reduce our national suicide rate by 20% within 5 years (2). One strategy proposed is to identify and target subgroups at greatest risk. Heuristic models outlining the chain of events leading to suicide often include biological or genetic characteristics, early life events including trauma and other stressors, impulsive aggressive traits, psychopathology, inadequate social support, and access to lethal means (3–5).
A growing body of evidence suggests that suicide vulnerability may be due to epigenetic alterations in molecular pathways important for hypothalamic-pituitary-adrenal (HPA) axis function. For example, DNA methylation changes in the NR3C1 gene, which encodes the glucocorticoid receptor, are altered by maternal behavior in rats (6) and are higher than normal in the hippocampus of suicide decedents who experienced early life trauma (7). The cumulative effect of these epigenetically mediated events is a reduction in glucocorticoid receptor levels, possibly leading to impaired responses to stressors. Suicidal individuals exhibit less ability to suppress cortisol after experimental administration of the synthetic glucocorticoid dexamethasone (8), and the cortisol stress response has been identified as one of the most promising candidate suicide endophenotypes (3). Other studies have provided evidence that first-degree relatives of suicide decedents fail to mount a proper HPA axis response to stress (9). Such findings are consistent with the diathesis-stress or dual-risk hypothesis, whereby an underlying biological state moderates an aberrant response to stress (10–14). Identification of the underlying genetic and epigenetic factors influencing vulnerability to suicidal behaviors in the context of stressors is needed to maximize suicide prevention efforts.
The objective of this study was to use genome-wide screening techniques to identify novel epigenetic associations in postmortem brain tissue of suicide decedents, followed by replication and functional assessment of identified loci. A secondary objective was to assess the degree to which identified loci would be present in peripheral blood samples and to evaluate their biomarker efficacy in the context of stress and anxiety.
Method
Human Samples
Postmortem prefrontal cortical tissue samples were obtained from the National Institute of Child Health and Human Development (NICHD) Brain and Tissue Bank for Developmental Disorders at the University of Maryland (20), the Stanley Medical Research Institute, and the Harvard Brain Bank at McLean Hospital. Peripheral blood was obtained from three Johns Hopkins studies of participants who consented to blood drawing for future research, including the Genetics of Recurrent Early-Onset Depression (GenRED) offspring (15, 16), the Prevention Research Center study participants (17, 18), and a prospective cohort of pregnant women described previously (19). Descriptions of the cohorts can be found in Table 1 and in Figure S1 and the supplementary methods in the data supplement accompanying the online version of this article.
| Sample Type, Cohort,a and Diagnostic Group | N | Suicidal Behavior | Age (years) | Sex | Substance Useb | Psychiatric Medication | Postmortem Interval (hours) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yes | No | Mean | SD | M | F | Yes | No | Yes | No | Mean | SD | ||
| Postmortem brain; suicide assessed | |||||||||||||
| NICHD | |||||||||||||
| Major depression | 29 | 21 | 8 | 32.0 | 15.9 | 14 | 15 | 9 | 20 | 12 | 17 | 18.10 | 7.1 |
| Control group | 29 | 2 | 27 | 32.1 | 16.1 | 14 | 15 | 2 | 27 | 0 | 29 | 16.14 | 5.0 |
| Stanley Medical Research Institute | |||||||||||||
| Bipolar disorder | 28 | 13 | 15 | 46.1 | 11.1 | 13 | 15 | 24 | 3 | 27 | 1 | 39.21 | 19.7 |
| Control group | 29 | 0 | 29 | 43.8 | 7.7 | 22 | 7 | 16 | 13 | 0 | 29 | 29.10 | 13.7 |
| Schizophrenia | 29 | 6 | 23 | 43.1 | 6.6 | 22 | 7 | 19 | 8 | 29 | 0 | 31.86 | 16.1 |
| McLean | |||||||||||||
| Bipolar disorder | 12 | 4 | 8 | 60.5 | 20.0 | 7 | 5 | 1 | 11 | 8 | 4 | 21.07 | 10.5 |
| Control group | 12 | 0 | 12 | 61.7 | 16.2 | 8 | 4 | 4 | 8 | 0 | 12 | 21.33 | 5.7 |
| Blood from live subjects | |||||||||||||
| GenRED offspring (15, 16) | |||||||||||||
| Bipolar disorder | 1 | 21 | 0 | 1 | 1 | 0 | 0 | 1 | |||||
| Suicidal ideation | 1 | 0 | |||||||||||
| Suicide attempt | 1 | 0 | |||||||||||
| Major depression | 8 | 18.1 | 3.0 | 6 | 2 | 2 | 6 | 1 | 7 | ||||
| Suicidal ideation | 3 | 5 | |||||||||||
| Suicide attempt | 2 | 6 | |||||||||||
| Control group | 13 | 15.3 | 2.3 | 5 | 8 | 0 | 13 | 4 | 9 | ||||
| Suicidal ideation | 3 | 10 | |||||||||||
| Suicide attempt | 1 | 12 | |||||||||||
| Prospective cohort of pregnant women (19); suicidal ideation assessed | |||||||||||||
| Bipolar disorder | 14 | 3 | 11 | 29.3 | 6.4 | 0 | 14 | 1 | 13 | 8 | 6 | ||
| Major depression | 37 | 10 | 27 | 31.5 | 6.4 | 0 | 37 | 1 | 36 | 25 | 12 | ||
| Prevention Research Center study (17, 18) | |||||||||||||
| Major depression | 30 | 29.6 | 1.2 | 11 | 19 | 17 | 13 | — | — | ||||
| Suicidal ideation | 20 | 10 | |||||||||||
| Suicide attempt | 15 | 15 | |||||||||||
| Control group | 295 | 30.5 | 2.6 | 117 | 178 | 70 | 225 | — | — | ||||
| Suicidal ideation | 59 | 236 | |||||||||||
| Suicide attempt | 33 | 262 | |||||||||||
TABLE 1. Characteristics of Subjects in Postmortem and Living Cohorts Examined for Epigenetic and Epigenetic Biomarkers of Suicide or Suicidal Behavior
Procedures
Genome-wide DNA methylation data were obtained from Illumina HM450 microarrays (Illumina, San Diego) previously generated by our group (20), for which data are located under Gene Expression Omnibus accession GSE15745. A discovery set of NICHD prefrontal cortical tissue data was created from 10 Caucasian individuals with major depression for whom bulk tissue data were available; seven died by suicide, and three did not. A replication set consisted of the remaining eight suicide and four nonsuicide samples from Caucasians with major depression in the NICHD cohort not originally interrogated.
Pyrosequencing was conducted in microarray-identified loci in all individuals in Table 1. All SKA2 gene expression data and rs7208505 genotyping was performed by using quantitative real-time polymerase chain reaction (PCR). Detailed methodological information, including salivary cortisol analysis for the GenRED offspring, is available in the supplementary methods and Table S1 in the online data supplement.
Statistical Analysis
Unless otherwise stated, the reported statistics derive from linear regression analysis, adjusted for age, sex, race, and postmortem interval (in brain cohorts), generated in R (http://www.r-project.org/). Relevant additional covariates were adjusted according to the strategy presented in the online supplementary methods information (Table S2). Using the Cramer–von Mises test, we subsequently evaluated all data distributions that rejected the null hypothesis of normality with nonparametric tests. All statistical tests were two tailed, with p≤0.05 denoting statistical significance. Microarray analysis employed false discovery rate correction for multiple testing. Where specified, genotype correction of SKA2 3′ untranslated region (UTR) DNA methylation was achieved by taking the residuals of a linear model of SKA2 3′ UTR DNA methylation as a function of rs7208505 genotype. Randomization was employed within all experimental processing batches. Personnel performing laboratory experiments were blind to caseness.
Results
Genome-Wide DNA Methylation Analysis and Replication
We performed a genome-wide screen for DNA methylation variation associated with suicide in a small discovery set of postmortem prefrontal cortical tissue from the NICHD cohort. Using a linear model adjusting for age and sex as covariates, we identified four loci significantly associated with suicide after correction for multiple testing, corresponding to the ATP8A1 (cg24533989), SKA2 (cg13989295), LOC153328 (cg15918259), and KCNAB2 (cg17106415) genes (Figure 1A). Using fluorescence-activated cell sorting, we separated neuronal and glial nuclei as described previously (20), after which only SKA2 exhibited nominal significance in the neuronal and glial fractions of both the discovery and replication sets (Figure 1B). The identified cytosine-guanine (CpG) dinucleotide is located on the antisense strand of chromosome 17 at position 57187729 (hg19) within the 3′ UTR of the SKA2 (spindle and kinetochore associated complex subunit 2) gene, which encodes a scaffold protein implicated in chaperoning the glucocordicoid receptor into the nucleus (21). It is important to note that the cytosine (C) at this position represents the alternative allele of single-nucleotide polymorphism (SNP) rs7208505, while the reference allele is a thymine (T). Also, the T allele abrogates the CpG dinucleotide and cannot be methylated. The assessment of rs7208505 epigenetic and genetic variation in an additive linear model demonstrated significant associations of both model terms with suicide across the entire NICHD cohort of 23 suicide cases and 35 comparison cases independent of ethnicity or DSM-IV diagnosis (Table 2). These associations were replicated in two independent cohorts of postmortem prefrontal cortical samples from the Stanley Medical Research Institute cohort and the Harvard Brain Bank at McLean Hospital cohort (Figure 1E, Table 2) and did not appear to be related to the mode of death (Result S1 in the supplementary results section of the data supplement accompanying the online version of this article).
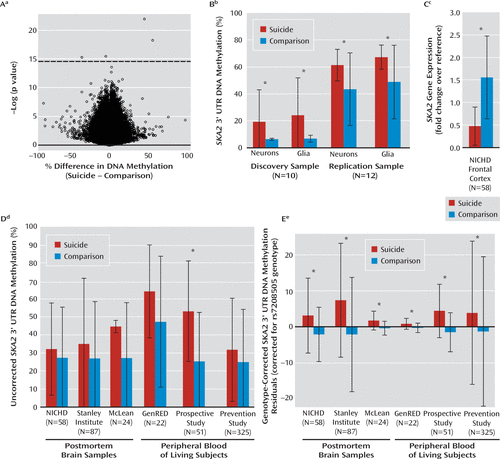
FIGURE 1. Discovery and Functional Analysis of SKA2 Gene As Epigenetic Biomarker of Suicide or Suicidal Behavior
a Volcano plot of DNA methylation differences, based on a linear model (x axis) and negative natural log of the p value (y axis), for postmortem brains from seven suicide and three nonsuicide subjects with major depression from the National Institute of Child Health and Development (NICHD) generated in bulk tissue. Loci remaining significant after false discovery rate correction appear above the dashed line, identifying significant hits at ATP8A1 (F=3424.5, df=3, 6, p<0.001), SKA2 (F=1101.7, df=3, 6, p<0.001), LOC153328 (F=384.3, df=3, 6, p<0.001), and KCNAB2 (F=243.5, df=3, 6, p<0.001).
b Significant differences in DNA methylation for the cg13989295 Illumina probe within the SKA2 3′ untranslated region (UTR) for the discovery sample neurons (F=784.3, df=3, 6, p<0.001), discovery sample glia (F=421.3, df=3, 6, p<0.001), replication sample neurons (F=2.2, df=3, 8, p=0.04), and replication sample glia (F=3.2, df=3, 8, p=0.02). Error bars represent standard deviations; large error bars derive from unaccounted-for variation in rs7208505 genotype.
c Significant differences in SKA2 gene expression in prefrontal cortical samples between 23 suicide and 35 comparison subjects from the NICHD cohort (Wilcoxon rank sum test: W=97, df=49.91, p<0.001). Error bars represent standard deviations.
d Uncorrected DNA methylation levels at rs7208505. Error bars represent standard deviations. Associated significance metrics are presented in Table S4 of the data supplement accompanying the online version of this article. NICHD, National Institute of Child Health and Human Development; Stanley Institute, Stanley Medical Research Institute; McLean, Harvard Brain Bank at McLean Hospital; GenRED, Genetics of Recurrent Early-Onset Depression (15, 16); Prospective Study, prospective cohort of pregnant women (19); Prevention Study, Prevention Research Center study (17, 18).
e Genotype-corrected DNA methylation levels at rs7208505. Error bars represent standard deviations. Associated significance metrics are located in Table 2. Abbreviations are provided in the preceding footnote.
* p≤0.05.
| Subjects | Model | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample Type, Cohort,a and Model Terms | N | Risk Variable | β | Error | p | F | df | R2 | p |
| Postmortem brain | |||||||||
| NICHD, neurons | 58 | Suicide | 2.2 | 8, 48 | 0.27 | <0.05 | |||
| DNA methylation | 0.03 | 0.01 | 0.03 | ||||||
| rs7208505 C/T | –0.58 | 0.27 | 0.04 | ||||||
| rs7208505 C/Cb | –1.50 | 0.57 | 0.04 | ||||||
| Age | –0.01 | 0.004 | 0.02 | ||||||
| Sex | –0.08 | 0.13 | 0.54 | ||||||
| Postmortem interval | 0.02 | 0.01 | 0.09 | ||||||
| Stanley Medical Research Institute | 86 | Suicide | 2.4 | 10, 75 | 0.24 | 0.02 | |||
| DNA methylation | 0.01 | 0.003 | 0.02 | ||||||
| rs7208505 C/T | –0.30 | 0.13 | 0.03 | ||||||
| rs7208505 C/Cb | –0.47 | 0.22 | 0.04 | ||||||
| Age | –0.00 | 0.01 | 0.44 | ||||||
| Sex | 0.17 | 0.09 | 0.07 | ||||||
| Postmortem interval | 0.004 | 0.003 | 0.17 | ||||||
| McLean | 24 | Suicide | 8.3 | 6, 17 | 0.75 | <0.001 | |||
| DNA methylation | 0.09 | 0.02 | 0.002 | ||||||
| rs7208505 C/T | –3.40 | 0.96 | 0.003 | ||||||
| rs7208505 C/Cb | –7.10 | 1.90 | 0.002 | ||||||
| Age | –0.01 | 0.003 | 0.0002 | ||||||
| Sex | 0.04 | 0.11 | 0.75 | ||||||
| Postmortem interval | –0.01 | 0.01 | 0.35 | ||||||
| Blood from live subjects | |||||||||
| GenRED offspring (15, 16) | 22 | Suicidal ideation | 2.0 | 6, 12 | 0.50 | 0.14 | |||
| DNA methylation | 0.17 | 0.07 | <0.05 | ||||||
| rs7208505 C/T | –6.50 | 3.10 | 0.06 | ||||||
| rs7208505 C/Cb | –15.00 | 6.70 | <0.05 | ||||||
| Age | 0.03 | 0.04 | 0.41 | ||||||
| Sex | –0.09 | 0.21 | 0.69 | ||||||
| Prospective cohort of pregnant women (19) | 51 | Suicidal ideation | 2.7 | 8, 42 | 0.34 | 0.02 | |||
| DNA methylation | 0.07 | 0.02 | <0.001 | ||||||
| rs7208505 C/T | –3.00 | 1.00 | 0.005 | ||||||
| rs7208505 C/Cb | –6.10 | 1.80 | 0.002 | ||||||
| Age | 0.02 | 0.02 | 0.41 | ||||||
| Sex | |||||||||
| Prevention Research Center study (17, 18) | 325 | Suicidal ideation | 3.9 | 10, 314 | 0.11 | <0.001 | |||
| DNA methylation | 0.002 | 0.001 | <0.05 | ||||||
| rs7208505 C/T | –0.05 | 0.06 | 0.40 | ||||||
| rs7208505 C/Cb | –0.14 | 0.10 | 0.16 | ||||||
| Age | 0.01 | 0.01 | 0.51 | ||||||
| Sex | –0.002 | 0.05 | 0.97 | ||||||
TABLE 2. Epigenetic and Genetic Effects of SKA2 rs7208505 on Suicide and Suicidal Behavior in Postmortem and Living Cohorts
Gene Expression and Functional Relevance of Identified Locus
In the NICHD brain cohort, SKA2 exhibited significantly lower gene expression values in the suicide cases than in the comparison cases (Figure 1C). SKA2 expression was significantly negatively associated with DNA methylation both before and after we controlled for rs7208505 variation, while genotype alone was not associated (Figure S2A and Table S3 in the online data supplement).
To understand the effects of 3′ UTR epigenetic variation on expression, we correlated rs7208505 variation with all other CpGs located across SKA2 by using available microarray data. Significant correlations were observed in neuronal but not glial DNA in two regions including promoter CpGs flanking a binding site for the CREB transcription factor and upstream of an intronic microRNA, miR-301a (online supplement: Figure S2 and Result S2). Epigenetic variation in these regions showed opposing effects on SKA2 gene expression (Figure S2 and Result S2). Average levels of DNA methylation of CpGs in the region upstream of miR-301a were lower in glial than in neuronal DNA, possibly accounting for the neuron-specific associations observed in this region (online supplement: Figure S2E). Together, rs7208505 epigenetic and genetic variation interacted with promoter and miR-301a proximal epigenetic variation to explain 40% of SKA2 gene expression, whereas 16% was explained by the model with rs7208505 variation alone (online supplement: Table S3).
Replication in Peripheral Tissues
We assessed the association of SKA2 variation with suicidal ideation in peripheral blood samples from the three living cohorts. Across all cohorts, significant rs7208505 DNA methylation elevations were observed, consistent with the brain findings (Figures 1D and 1E, Table 2, online Table S4, online Result S3). Model factors were significant among the 30 women in the prospective cohort who provided blood samples months before the suicidal ideation measurement (online supplement: Table S5, Result S4), suggesting that SKA2 3′ UTR DNA methylation variation (relevant to suicidal ideation) preceded the transition to suicidal ideation. Weighted gene coexpression network analysis (22) in brain- and blood-derived data provided supporting evidence that peripheral epigenetic variation is a marker of primarily neuronal processes (online supplement: Result S5).
Association of SKA2 With Salivary Cortisol
Using prospectively recorded cortisol measurements in the GenRED offspring cohort, we assessed the ability of SKA2 3′ UTR epigenetic and genetic variation to mediate suppression of cortisol levels. Waking cortisol level was significantly associated with suicidal ideation in this cohort; however, cortisol measured at 30 or 60 minutes was not associated, nor were CpGs in the region directly upstream of rs7208505 (online supplement: Figure S4A, Result S6). Only waking cortisol was significantly associated with epigenetic variation at rs7208505 (online supplement: Table S6, Figure S4B); however, as SKA2 is implicated in glucocorticoid signaling, we reasoned that cortisol levels may interact with SKA2 to mediate suppression of future cortisol. Prospectively investigated interaction of waking cortisol with SKA2 3′ UTR epigenetic and genetic variation was significantly associated with the reduced suppression of cortisol from the 30 to 60 minute time points (online Table S6). SKA2-mediated changes in glucocorticoid signaling may influence and interact with other suicidal-ideation-related biological variation, such as promoter CpGs in the SAT1 gene (online supplement: Result S7 and Figures S4C and S4D) where gene expression has been previously implicated in suicidal behavior (23).
Interaction of SKA2 With Anxiety and Stress
We observed a significant interaction of both perceived stress scores and anxiety scores with rs7208505 genotype and DNA methylation in predicting third-trimester suicidal ideation from first- and second-trimester blood in the prospective cohort (Table 3, online Figure S5). This association was replicated in the GenRED offspring cohort, where anxiety interacted with rs7208505 epigenetic and genetic status to moderate suicide attempt but not suicidal ideation (Table 3). In the Prevention Research Center cohort, the anxiety interaction model was associated with suicidal ideation; however, the association became even stronger in the subset with suicide attempts (Table 3, online Figure S5). The interaction of SKA2 3′ UTR DNA methylation with anxiety had a nearly significant association with suicide attempt in those with suicidal ideation (online Table S6). Finally, a nonsignificant interaction between DNA methylation and anxiety was observed with the metric “intent to die” among individuals who had attempted suicide (Table S6). Cumulatively, our data suggest that epigenetic variation at SKA2 could increase risk for suicidal ideation and that among individuals with suicidal ideation also experiencing anxiety or stress, a suicide attempt is more likely.
| Subjects | Model | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cohort and Model Termsa | N | Risk Variable | β | Error | p | F | df | R2 | p |
| Prospective cohort of pregnant women (19) | 30 | Suicidal ideation | 6.8 | 10, 17 | 0.80 | <0.001 | |||
| DNA methylation | –0.07 | 0.05 | 0.14 | ||||||
| rs7208505 C/T | 2.66 | 2.32 | 0.27 | ||||||
| rs7208505 C/Cb | 6.30 | 4.42 | 0.17 | ||||||
| Anxiety | –0.13 | 0.13 | 0.31 | ||||||
| Age | 0.01 | 0.01 | 0.48 | ||||||
| Sex | |||||||||
| Interaction | |||||||||
| DNA methylation with anxiety | 0.05 | 0.02 | 0.007 | ||||||
| C/T genotype with anxiety | –1.92 | 0.78 | 0.03 | ||||||
| C/C genotype with anxiety | –4.18 | 1.46 | 0.02 | ||||||
| GenRED offspring (15, 16) | 22 | Suicide attempt | 4.3 | 9, 12 | 0.80 | 0.02 | |||
| DNA methylation | 0.01 | 0.04 | 0.89 | ||||||
| rs7208505 C/T | –0.23 | 1.60 | 0.89 | ||||||
| rs7208505 C/Cb | –0.34 | 3.50 | 0.92 | ||||||
| Anxiety | –0.32 | 0.30 | 0.30 | ||||||
| Age | 0.04 | 0.03 | 0.15 | ||||||
| Sex | 0.21 | 0.13 | 0.14 | ||||||
| Interaction | |||||||||
| DNA methylation with anxiety | 0.21 | 0.08 | 0.02 | ||||||
| C/T genotype with anxiety | –7.80 | 3.20 | 0.04 | ||||||
| C/C genotype with anxiety | –18.00 | 7.10 | 0.03 | ||||||
| Prevention Research Center study (17, 18) | 325 | Suicidal ideation | 2.6 | 12, 312 | 0.09 | 0.003 | |||
| DNA methylation | 0.001 | 0.001 | 0.37 | ||||||
| rs7208505 C/T | –0.02 | 0.06 | 0.79 | ||||||
| rs7208505 C/Cb | –0.05 | 0.11 | 0.66 | ||||||
| Anxiety | 0.15 | 0.15 | 0.32 | ||||||
| Age | 0.004 | 0.01 | 0.66 | ||||||
| Sex | –0.03 | 0.05 | 0.58 | ||||||
| Interaction | |||||||||
| DNA methylation with anxiety | 0.01 | 0.004 | 0.04 | ||||||
| C/T genotype with anxiety | –0.39 | 0.24 | 0.10 | ||||||
| C/C genotype with anxiety | –0.98 | 0.41 | 0.02 | ||||||
| Prevention Research Center study (17, 18) | 325 | Suicide attempt | 1.9 | 12, 312 | 0.07 | 0.04 | |||
| DNA methylation | 0.0004 | 0.001 | 0.70 | ||||||
| rs7208505 C/T | –0.01 | 0.05 | 0.79 | ||||||
| rs7208505 C/Cb | 0.05 | 0.09 | 0.54 | ||||||
| Anxiety | –0.11 | 0.12 | 0.36 | ||||||
| Age | 0.002 | 0.01 | 0.83 | ||||||
| Sex | –0.01 | 0.04 | 0.87 | ||||||
| Interaction | |||||||||
| DNA methylation with anxiety | 0.01 | 0.004 | 0.004 | ||||||
| C/T genotype with anxiety | –0.11 | 0.20 | 0.57 | ||||||
| C/C genotype with anxiety | –0.95 | 0.34 | 0.006 | ||||||
TABLE 3. Effects on Suicidal Behavior of Interactions Between Anxiety and SKA2 rs7208505 in Three Living Cohorts
Prediction of Suicidal Behavior
We assessed the ability of our statistical model to predict suicidal ideation in peripheral tissues of living individuals. We used suicide attempt data from the Prevention Research Center cohort to generate an additive linear model of rs7208505 genotype and SKA2 3′ UTR DNA methylation interacting with anxiety status, controlling for age and sex as covariates. With anxiety status used as the interactive covariate, the model predicted suicidal ideation in the GenRED cohort with an area under the receiver operator characteristic curve (AUC) of 0.71 (95% CI=0.42–1.00); however, use of salivary cortisol level as the interactive covariate improved the AUC to 0.82 (95% CI=0.60–1.00) (Figure 2A). In the prospective sample, the perceived stress metric at the time of blood drawing was used as the interactive covariate, resulting in an AUC for prediction of suicidal ideation of 0.80 (95% CI=0.64–0.97) (Figure 2A). Limiting the women in the prospective study to the 30 for whom third-trimester suicidal ideation was predicted from first- or second-trimester blood generated an AUC of 0.79 (95% CI=0.42–1.00) (Figure 2B). Increasing the stringency of the threshold to define suicidal ideation (see supplementary methods in the online data supplement) resulted in improved model performance across both comparisons in this cohort (all women: N=51, AUC=0.91, 95% CI=0.8–1.00; first- or second-trimester women: N=30, AUC=0.96, 95% CI=0.89–1.00). In the GenRED cohort, the model predicted the four suicide attempters from the group with an AUC of 0.97 (95% CI=0.89–1.00) (Figure 2B).
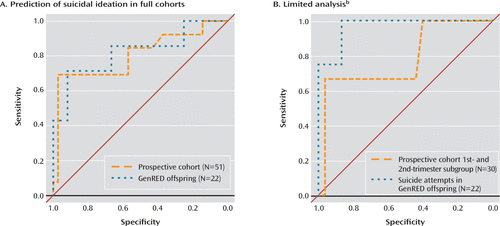
FIGURE 2. Prediction of Suicidal Behavior by Interaction of Anxiety With SKA2 3′ UTR DNA Methylation and SKA2 rs7208505 Genotype in Two Living Cohortsa
a Data on suicide attempts from the Prevention Research Center study (17, 18) were used to generate an additive linear model of the interaction of anxiety status with SKA2 3′ untranslated region (UTR) DNA methylation and rs7208505 genotype, with age and sex covariates controlled for. Receiver operator characteristic (ROC) curves depict the sensitivity as a function of the specificity for predictions of suicidal ideation or behavior. GenRED, Genetics of Recurrent Early-Onset Depression (15, 16); prospective cohort, prospective cohort of pregnant women (19). Definitions of anxiety are given in the “Study metrics” section of the data supplement accompanying the online version of this article.
b ROC curves are shown for suicide attempts in the GenRED offspring cohort (N=4) and for women in the prospective cohort for whom suicidal ideation in the third trimester was predicted from blood sampled in the first or second trimester.
Discussion
Using microarray technology to scan for epigenetic suicide associations, we identified a significant effect in a very small group of suicide decedents. The effect size of about 55% DNA methylation difference at SKA2 enabled this small study group to have adequate power to survive correction for multiple testing, which was driven by the underlying genetic status of the rs7208505 SNP, which abrogates the CpG dinucleotide. While microarray analysis was performed only in Caucasians, incorporation of both the genetic and epigenetic variation at this locus enabled replication across the entirety of the NICHD cohort, two additional postmortem brain cohorts, and three blood cohorts. Despite the consistency of the findings, the relatively small sizes of the studied cohorts suggest they represent promising but preliminary results warranting further study. The presented linear models suggested that DNA methylation and rs7208505 genotype may have opposing effects on suicidal behavior; however, as these metrics were highly correlated, the protective effects of rs7208505 may represent a statistical artifact. Analysis of genetic and epigenetic effects on suicidal behavior and gene expression separately indicated that DNA methylation alone may be the primary factor conferring risk. It is important that the overall proportion of DNA methylation at rs720505 increased significantly with each successive C-containing allele, suggesting that the underlying genetic architecture at rs7208505 may confer vulnerability by providing a genetic template for methylation changes to occur. This risk template would be expected to vary as a function of ethnicity, as allele frequencies for the C-containing allele of rs7208505 are reportedly much lower in African Americans (about 18%) than in other ethnicities (50%−60%). Cumulatively, numerous consistent associations were observed with suicidal ideation, suicide attempt, and suicide completion, independent of variation in ethnicity and psychiatric diagnosis, suggesting that variation in SKA2 may mediate risk for suicidal behaviors that progress from ideation to attempt to suicide.
SKA2 may influence suicidal phenotypes through its role in chaperoning the glucocorticoid receptor from the cytoplasm to the nucleus. Rice et al. (21) demonstrated that knockdown of SKA2 eliminated glucocorticoid receptor transactivation and response to dexamethasone treatment in vitro and that protein levels of SKA2 were decreased by glucocorticoid treatment, suggesting that SKA2 gene expression may be a component of the glucocorticoid feedback inhibition system. In our data, SKA2 genetic and epigenetic differences were associated with reduced suppression of salivary cortisol after waking in the GenRED cohort. As the blood was not drawn at the same time as the salivary cortisol sampling, the causative role of DNA methylation must be interpreted cautiously. While DNA methylation variation at rs7208505 might be important for suicidal ideation etiology, it remains possible that this variation is a reflection of cortisol variation.
In the proposed model, SKA2 epigenetic and genetic variation represents an underlying state increasing suicide risk in the presence of a stressor. SKA2 epigenetic and genetic variation interacted with stress and anxiety metrics to mediate suicidal ideation in the prospective cohort, while in the Prevention Research Center and GenRED offspring cohorts, the same model distinguished individuals with suicidal ideation who transitioned to suicide attempt. It is important to note that the suicidal ideation, suicide attempt, and suicide phenotypes are not interchangeable; however, in some individuals they represent progressive stages of suicidal behavior that share many etiological factors. The proportion of variance accounted for by our models was very high in some cohorts and leaves little room for the involvement of other factors. While our data suggest that SKA2 may be etiologically relevant to glucocorticoid signaling, it is possible that the detected epigenetic variation at SKA2 also represents a molecular record of suicide-dysregulated glucocorticoid load over time and thus may be reflective of other sources of etiologically relevant variation at other loci implicated in suicide. In a recent study, blood gene expression at SAT1 prospectively predicted both suicidal ideation and suicide attempt (23). Our supplemental analysis demonstrated an interaction between SKA2 variation and DNA methylation at a CpG in the SAT1 promoter located within a region enriched for glucocorticoid receptor binding. SKA2-mediated failure to suppress normal stress response may play a role in SAT1 gene expression variation and could contribute to the transition from suicidal ideation to suicide attempt. Cumulatively, our data are consistent with findings from an epidemiological study of 108,664 individuals in 21 countries that found disorders characterized by anxiety and poor impulse control predicted the transition from suicidal ideation to suicide attempt (24).
One caveat with these analyses is that different metrics of suicidal ideation, suicide attempt, stress, and anxiety were available across the studied cohorts. In the Prevention Research Center and prospective study groups, anxiety was measured by response to a single question, while the Screen for Child Anxiety Related Disorders (SCARED) was used for the GenRED offspring. In the prospective cohort, we showed that anxiety, as measured by the Edinburgh Postnatal Depression Scale, was highly associated with perceived stress, which in another study was correlated with salivary cortisol level (25). Thus, it is possible that the observed interactions of SKA2 with anxiety across cohorts are a reflection of underlying differences in stress and HPA axis response in anxious individuals. Despite these limitations, the ability of the linear model generated from the Prevention Research Center cohort to accurately predict both suicidal ideation and suicide attempt in the prospective and GenRED cohorts suggests that consistency was captured by these diverse metrics.
The lower SKA2 gene expression in NICHD brains from suicide decedents was associated primarily with isolated neuronal nuclei, suggesting the epigenetic dysregulation may be confined to neurons. The postmortem brain data were generated in the prefrontal cortex, a brain region with inhibitory connections to the HPA axis (26, 27) and responsible for decision making, inhibition of negative thoughts, and impulsivity (28, 29). Low glucocorticoid receptor transactivation is consistent with current models of suicide diathesis, as experimentally reduced glucocorticoid receptor gene expression in rodents mimics suicidal human characteristics (8), producing increases in corticosterone and helplessness in response to stress (30).
The influence of SKA2 3′ UTR epigenetic variation on gene expression appeared to be mediated by interaction with epigenetic variation within the gene promoter and proximal to intronic miR-301a, which has previously been shown to be low in the postmortem prefrontal cortex of individuals who die by suicide (31). Critically, miR-301a modulates SKA2 gene expression in A549 cell models by indirectly inhibiting CREB binding to the SKA2 promoter (32), while the promoter CpGs shown to correlate with rs7208505 DNA methylation directly flank this CREB binding site. As would be expected given the model, we observed a significant effect of the interaction of miR-301a and promoter CpG variation on SKA2 expression. The observed correlations of SKA2 3′ UTR DNA methylation with other CpGs across the gene could result from common epigenetic reprogramming effects of glucocorticoid receptor binding, as the regions demonstrating correlations were located within glucocorticoid receptor immunoprecipitation peaks identified from Encyclopedia of DNA Elements (ENCODE) data. It is also possible that miR-301a proximal genetic variation in linkage disequilibrium with rs7208505 serves to alter glucocorticoid receptor recruitment, subsequently reprogramming DNA methylation in the region, as discussed in the online supplementary material. The miR-301a is an intronic microRNA and requires mRNA transcription of SKA2 to be generated by Drosha, an RNase III enzyme (33). DNA methylation upstream of miR-301a may therefore result in cotranscriptional slowing and allow for spliceosomal interaction as occurs with inclusion of methylated alternative exons in alternatively spliced genes (34). Elevated neuronal but not glial DNA methylation levels proximal to miR-301a suggest a possible functionally different effect of miR-301a in these two cell types. It is important to note that while epigenetic variation proximal to the miR-301a and CREB binding site was associated with SKA2 gene expression, it was not associated with suicidal phenotype.
While a growing number of studies are investigating epigenetic alterations in suicide (23, 35–40), few studies have identified biomarkers with high prediction accuracy. To our knowledge, the biomarker identified in this study represents the first genetic and epigenetic biomarker capable of predicting suicidal ideation and suicide attempt in a prospective manner with over 80% accuracy from blood. The model performed remarkably well at predicting suicide attempt in the GenRED cohort; however, with only four attempters, this result should be interpreted with caution. While the Prevention Research Center cohort contained many more suicide attempt cases, we did not test prediction in this cohort as the time between suicide attempt and blood draw was greater than 10 years on average. Accumulating epigenetic change due to stochastic drift, substance use, and errors in retrospective reporting would call into question the reliability of the prediction. However, this highlights the fact that the cause versus effect of prediction accuracy in the GenRED offspring cohort must also be interpreted with care as the blood was taken after the suicide attempt. Nevertheless, our data demonstrate similar accuracies when predicting suicidal ideation in a prospective manner, suggesting that SKA2 epigenetic and genetic variation may represent a trait influencing underlying suicide risk when interacting with stress. Cumulatively, the clinical implications of this finding are that early screening of those at risk for suicidal ideation and suicide attempt may be possible, allowing for the identification of individuals at risk, proactive treatment, and stress and anxiety reduction. The potential efficacy of this biomarker is relevant to numerous populations, for example, the military, where the identification of an underlying vulnerability may identify individuals at risk for developing suicidal behaviors when exposed to the stress of war-time situations. Future studies should be carried out to further evaluate the prospective efficacy of this finding in additional populations.
1
2 : A strategic approach for prioritizing research and action to prevent suicide. Psychiatr Serv 2013; 64:71–75Link, Google Scholar
3 : Candidate endophenotypes for genetic studies of suicidal behavior. Biol Psychiatry 2009; 65:556–563Crossref, Medline, Google Scholar
4 : The relationship of impulsive aggressiveness to suicidality and other depression-linked behaviors. Curr Psychiatry Rep 2007; 9:460–466Crossref, Medline, Google Scholar
5 : Methods of adolescent suicide prevention. J Clin Psychiatry 1999; 60(suppl 2):70–74Medline, Google Scholar
6 : Maternal programming of individual differences in defensive responses in the rat. Ann N Y Acad Sci 2004; 1032:85–103Crossref, Medline, Google Scholar
7 : Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci 2009; 12:342–348Crossref, Medline, Google Scholar
8 : The dexamethasone suppression test and suicide prediction. Am J Psychiatry 2001; 158:748–753Link, Google Scholar
9 : Dysregulation of the sympathetic nervous system, hypothalamic-pituitary-adrenal axis and executive function in individuals at risk for suicide. J Psychiatry Neurosci 2010; 35:399–408Crossref, Medline, Google Scholar
10 : Toward a clinical model of suicidal behavior in psychiatric patients. Am J Psychiatry 1999; 156:181–189Link, Google Scholar
11 : Beyond diathesis stress: differential susceptibility to environmental influences. Psychol Bull 2009; 135:885–908Crossref, Medline, Google Scholar
12 : Biological sensitivity to context: the interactive effects of stress reactivity and family adversity on socioemotional behavior and school readiness. Child Dev 2010; 81:270–289Crossref, Medline, Google Scholar
13 : Biological sensitivity to context, I: an evolutionary-developmental theory of the origins and functions of stress reactivity. Dev Psychopathol 2005; 17:271–301Crossref, Medline, Google Scholar
14 : Developmental systems: contexts and evolution, in Handbook of Child Psychology, Vol 1. Edited by Mussen P. New York, Wiley, 1983, pp 237–294Google Scholar
15 : Genetics of Recurrent Early-Onset Depression (GenRED): design and preliminary clinical characteristics of a repository sample for genetic linkage studies. Am J Med Genet B Neuropsychiatr Genet 2003; 119B:118–130Crossref, Medline, Google Scholar
16 : Genome-wide association study of recurrent early-onset major depressive disorder. Mol Psychiatry 2011; 16:193–201Crossref, Medline, Google Scholar
17 : Developmental epidemiologically based preventive trials: baseline modeling of early target behaviors and depressive symptoms. Am J Community Psychol 1991; 19:563–584Crossref, Medline, Google Scholar
18 : The course and malleability of aggressive behavior from early first grade into middle school: results of a developmental epidemiologically-based preventive trial. J Child Psychol Psychiatry 1994; 35:259–281Crossref, Medline, Google Scholar
19 : Antenatal prediction of postpartum depression with blood DNA methylation biomarkers. Mol Psychiatry 2014; 19:560–567; correction, 2014; 19:633Crossref, Medline, Google Scholar
20 : A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics 2013; 8:290–302Crossref, Medline, Google Scholar
21 : Identification and functional analysis of SKA2 interaction with the glucocorticoid receptor. J Endocrinol 2008; 198:499–509Crossref, Medline, Google Scholar
22 : WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008; 9:559Crossref, Medline, Google Scholar
23 : Discovery and validation of blood biomarkers for suicidality. Mol Psychiatry 2013; 18:1249–1264Crossref, Medline, Google Scholar
24 : Cross-national analysis of the associations among mental disorders and suicidal behavior: findings from the WHO World Mental Health Surveys. PLoS Med 2009; 6:e1000123Crossref, Medline, Google Scholar
25 : Effect of the emotional freedom technique on perceived stress, quality of life, and cortisol salivary levels in tension-type headache sufferers: a randomized controlled trial. Explore (NY) 2013; 9:91–99Crossref, Medline, Google Scholar
26 ;
27 : The neurodevelopmental origins of suicidal behavior. Trends Neurosci 2012; 35:14–23Crossref, Medline, Google Scholar
28 : Human and rodent homologies in action control: corticostriatal determinants of goal-directed and habitual action. Neuropsychopharmacology 2010; 35:48–69Crossref, Medline, Google Scholar
29 : The role of the ventral medial prefrontal cortex in social decision making. J Neurosci 2009; 29:7631–7632Crossref, Medline, Google Scholar
30 : Mice with genetically altered glucocorticoid receptor expression show altered sensitivity for stress-induced depressive reactions. J Neurosci 2005; 25:6243–6250Crossref, Medline, Google Scholar
31 : MicroRNA expression is down-regulated and reorganized in prefrontal cortex of depressed suicide subjects. PLoS ONE 2012; 7:e33201Crossref, Medline, Google Scholar
32 : Intronic miR-301 feedback regulates its host gene, ska2, in A549 cells by targeting MEOX2 to affect ERK/CREB pathways. Biochem Biophys Res Commun 2010; 396:978–982Crossref, Medline, Google Scholar
33 : Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol 2008; 15:902–909Crossref, Medline, Google Scholar
34 : Contrasting chromatin organization of CpG islands and exons in the human genome. Genome Biol 2010; 11:R70Crossref, Medline, Google Scholar
35 : Analysis of CpG SNPs in 34 genes: association test with suicide attempt in schizophrenia. Schizophr Res 2013; 147:262–268Crossref, Medline, Google Scholar
36 : TrkB gene expression and DNA methylation state in Wernicke area does not associate with suicidal behavior. J Affect Disord 2011; 135:400–404Crossref, Medline, Google Scholar
37 : Genome-wide methylation changes in the brains of suicide completers. Am J Psychiatry 2013; 170:511–520Link, Google Scholar
38 : Differential glucocorticoid receptor exon 1(B), 1(C), and 1(H) expression and methylation in suicide completers with a history of childhood abuse. Biol Psychiatry 2012; 72:41–48Crossref, Medline, Google Scholar
39 : Genetic variation in DNMT3B and increased global DNA methylation is associated with suicide attempts in psychiatric patients. Genes Brain Behav 2013; 12:125–132Crossref, Medline, Google Scholar
40 : Epigenetic regulation of spermidine/spermine N1-acetyltransferase (SAT1) in suicide. J Psychiatr Res 2011; 45:1229–1235Crossref, Medline, Google Scholar



