Cross-Disorder Genome-Wide Analyses Suggest a Complex Genetic Relationship Between Tourette’s Syndrome and OCD
Abstract
Objective:
Obsessive-compulsive disorder (OCD) and Tourette’s syndrome are highly heritable neurodevelopmental disorders that are thought to share genetic risk factors. However, the identification of definitive susceptibility genes for these etiologically complex disorders remains elusive. The authors report a combined genome-wide association study (GWAS) of Tourette’s syndrome and OCD.
Method:
The authors conducted a GWAS in 2,723 cases (1,310 with OCD, 834 with Tourette’s syndrome, 579 with OCD plus Tourette’s syndrome/chronic tics), 5,667 ancestry-matched controls, and 290 OCD parent-child trios. GWAS summary statistics were examined for enrichment of functional variants associated with gene expression levels in brain regions. Polygenic score analyses were conducted to investigate the genetic architecture within and across the two disorders.
Results:
Although no individual single-nucleotide polymorphisms (SNPs) achieved genome-wide significance, the GWAS signals were enriched for SNPs strongly associated with variations in brain gene expression levels (expression quantitative loci, or eQTLs), suggesting the presence of true functional variants that contribute to risk of these disorders. Polygenic score analyses identified a significant polygenic component for OCD (p=2×10−4), predicting 3.2% of the phenotypic variance in an independent data set. In contrast, Tourette’s syndrome had a smaller, nonsignificant polygenic component, predicting only 0.6% of the phenotypic variance (p=0.06). No significant polygenic signal was detected across the two disorders, although the sample is likely underpowered to detect a modest shared signal. Furthermore, the OCD polygenic signal was significantly attenuated when cases with both OCD and co-occurring Tourette’s syndrome/chronic tics were included in the analysis (p=0.01).
Conclusions:
Previous work has shown that Tourette’s syndrome and OCD have some degree of shared genetic variation. However, the data from this study suggest that there are also distinct components to the genetic architectures of these two disorders. Furthermore, OCD with co-occurring Tourette’s syndrome/chronic tics may have different underlying genetic susceptibility compared with OCD alone.
Obsessive-compulsive disorder (OCD) [MIM 164230] and Tourette’s syndrome [MIM 137580] are highly familial neuropsychiatric disorders with complex overlapping genetic etiologies (1–3). Some 20%–60% of individuals with Tourette’s syndrome have co-occurring OCD, and 10%–20% of patients initially diagnosed with OCD have Tourette’s syndrome or chronic tics, rates well over what is expected based on their respective population prevalences (4–6). Both disorders are characterized by the presence of repetitive, ritualized, or stereotyped behaviors (tics and compulsions), often preceded by cognitive or sensory phenomena (premonitory urges and obsessions), and clinical differentiation of compulsions versus complex tics can be challenging (7). Genetic epidemiological studies suggest up to 90% shared genetic variance between Tourette’s syndrome/chronic tics and OCD (8–10), and abnormalities in cortico-striatal-thalamo-cortical circuitry have been identified in both conditions (1).
To date, most of the work aimed at elucidating the genetic causes of Tourette’s syndrome and OCD has focused on candidate gene studies and linkage analyses; a few studies examining chromosome abnormalities and copy number variants have also been reported (11–14). Recently, our group performed genome-wide association studies (GWAS) of Tourette’s syndrome and OCD, and for each disorder identified a number of genes and genomic regions of interest, most with modest significance levels. Here we report GWAS results for a combined sample of individuals with Tourette’s syndrome, OCD, or Tourette’s syndrome plus OCD, along with analyses aimed at elucidating the genetic architectures and genetic relationships between the two disorders. Combining these heterogeneous but related phenotypes in joint analyses could have one of two potential effects: 1) enhancement of the genetic signal as a consequence of increased power by adding samples from genetically related phenotypes; or 2) reduction of the genetic signal as a consequence of increased genetic heterogeneity, outweighing the potential benefits of increased sample size. Either way, given previous evidence supporting shared genetic factors and the lack of definitive susceptibility genes for either disorder, joint analyses of Tourette’s syndrome and OCD cases represent an important step toward understanding the underlying causes of these common neuropsychiatric disorders.
Method
Cases
Individuals with Tourette’s syndrome or OCD were recruited as part of collaborative efforts to conduct the first GWAS for these disorders (for details, see references 15, 16). Although data were collected independently for Tourette’s syndrome and OCD, all genotyping and data cleaning were done together, facilitating joint analyses. Participants who were age 18 or older provided written, voluntary informed consent; those under age 18 provided assent, and written parental consent for their participation was obtained. The study was approved by the ethics committees of all participating sites and was in accordance with the Declaration of Helsinki. For the cross-disorder analyses, any participant with either Tourette’s syndrome or OCD was considered affected. For details on the inclusion and exclusion criteria and assessment protocols, see the Supplementary Methods section in the data supplement that accompanies the online edition of this article.
Tourette’s syndrome.
The Tourette’s syndrome sample consisted of 1,286 individuals recruited from 20 sites in the United States, Canada, the United Kingdom, the Netherlands, and Israel, and included individuals of general European (EU) ancestry as well as two EU-derived population isolates of Ashkenazi Jewish (AJ) and French Canadian (FC) descent. Co-occurring OCD symptoms were assessed in 77% of participants; 46% of those evaluated had co-occurring OCD (N=452). OCD status was unknown for 300 individuals with Tourette’s syndrome.
OCD.
The OCD sample consisted of 1,437 individuals with OCD and 290 parent-child trios. While the original GWAS sample consisted of 1,865 OCD probands recruited from 21 sites in North, Central, and South America, Europe, the United Arab Emirates, and South Africa, only individuals of European ancestry (EU, AJ, and EU-derived Afrikaner [SA] descent) were included in the present study (16). Co-occurring Tourette’s syndrome or chronic tics was assessed in 77% of OCD probands; of these, 12% had comorbid Tourette’s syndrome or chronic tics (N=159). Tourette’s syndrome/chronic tics status was unknown for 405 OCD-affected individuals.
Controls
The EU control sample consisted of 4,975 European Caucasian controls primarily derived from cohorts of previously genotyped, unselected population controls (see Supplementary Methods in the online data supplement). Ancestry-matched controls for the FC (N=196) and SA (N=158) samples were collected in parallel with their respective cases (15, 16). Ancestry-matched controls for individuals with AJ ancestry were identified from the EU control sample based on self-reported ancestry and principal-components analysis (N=338).
Genotyping and Quality Control
Genotyping and quality control procedures have been described previously (15, 16; see also Supplementary Methods in the online data supplement). Briefly, case subjects and trios with Tourette’s syndrome or OCD and controls were randomized across plates and genotyped on the Illumina HumanHap610 SNP array (Illumina, San Diego) at the Broad Institute of Harvard-MIT (Cambridge, Mass.) or on the Illumina HumanHap370 at the Yale Center for Genome Analysis (New Haven, Conn.) (see Figure S1 in the online data supplement). Eighty-eight samples were genotyped on both platforms to allow for cross-platform concordance checks. Quality control analyses were performed using PLINK, version 1.07 (17) and EIGENSTRAT (18). Multidimensional scaling analysis was used to exclude case-control samples of non-European descent. Remaining EU and European-derived isolate samples were separated into four strata (EU, AJ, FC, and SA) based initially on self-reported ancestry and then on observed genetic ancestry (15, 16). Imputation was performed with 1000 Genomes Project data (June 2011 Data Release) (19) as the reference panel, using IMPUTE version 2.1.2 (20) (see Supplementary Methods).
Genome-Wide Association Analyses
Individual ancestry-stratified case-control genome-wide association analyses (EU, AJ, FC, and SA) and one case/pseudo-control analysis using the OCD trios were performed in PLINK using logistic regression under an additive model with significant subpopulation-specific multidimensional scaling axes included as covariates to control for residual population stratification (see Figure S2 in the online data supplement). These population-specific analyses were then combined in a fixed-effects model meta-analysis using case-weighting in METAL (21). Single-nucleotide polymorphisms (SNPs) with p values <10−5 were annotated with details including their genomic region and location, allele frequencies, nearby genes, and p values from individual Tourette’s syndrome and OCD GWAS studies. Heterogeneity tests were also conducted to assess subpopulation differences using Cochran’s Q and I2 statistics. As is standard in GWAS for complex traits, a genome-wide threshold of p<5×10−8 was considered statistically significant evidence of association (22, 23).
Enrichment Analyses
GWAS results were examined for enrichment of functional SNPs previously associated with gene expression levels in several brain regions (i.e., expression quantitative trait locus SNPs, eQTLs) or with variation in gene methylation levels (methylation QTLs, mQTLs). eQTL data were generated from cerebellum, parietal cortex, and frontal cortex (see Supplementary Methods in the online data supplement). mQTLs were derived from adult cerebellum (24). Only GWAS SNPs meeting high stringency criteria for eQTLs or mQTLs (p<10−6) were considered. For each phenotype (Tourette’s syndrome, OCD, combined), a quantile-quantile (Q-Q) plot of GWAS disease association p values was generated for eQTL and mQTL SNPs and compared with a standard Q-Q plot of GWAS p values expected under the null distribution assuming no enrichment. A leftward shift in the eQTL/mQTL Q-Q plot relative to the diagonal line (representing the null distribution) indicates enrichment of eQTLs/mQTLs. The level of enrichment of eQTLs or mQTLs in each brain tissue associated with Tourette’s syndrome or OCD was then quantified using a false discovery rate of <0.25, that is, 75% of observed SNPs represent true disease associations (see Supplementary Methods).
Polygenic Score Analysis
Polygenic score analyses were conducted in PLINK using genotyped SNPs to test the hypothesis that multiple genes of small effect jointly contribute to Tourette’s syndrome and OCD susceptibility and to explore the genetic relationships between these disorders (25). Samples were divided into nonoverlapping discovery and target samples (see Supplementary Methods in the online data supplement). For the primary OCD polygenic analysis, cases were restricted to subjects without known co-occurring Tourette’s syndrome/chronic tics. SNPs with GWAS p values passing predetermined significance thresholds (p<0.01, 0.1, 0.2, 0.3, 0.4, and 0.5) in the discovery sample were extracted along with their risk alleles and odds ratios, and then linkage disequilibrium (LD) pruned (r2<0.5). For each significance threshold, a quantitative aggregate risk score was calculated for each individual in the target sample, defined as the sum of the number of risk alleles present at each locus weighted by the log of the odds ratio for that locus estimated from the discovery sample. The relationship between aggregate risk score and case-control status in the target sample was examined at each significance threshold using logistic regression. The percentage of phenotypic variance explained by the aggregate risk score (Nagelkerke’s pseudo-R2) was estimated.
Two separate statistical approaches were used to determine the significance of the observed differences in polygenic risk score prediction between discovery samples. First, permutation testing was conducted to derive an empirical significance of the magnitude of change in R2 between polygenic risk scores derived from the discovery sample of Tourette’s syndrome/chronic tics without OCD compared with those derived from the “all OCD” sample (OCD with or without Tourette’s syndrome/chronic tics). Second, risk alleles from each discovery sample were used to calculate the difference in polygenic risk scores between the transmitted (case) alleles and the untransmitted (pseudo-control) alleles in the OCD trio target sample. The degree of risk score elevation (risk scoretransmitted − risk scoreuntransmitted) was then standardized ([risk scoretransmitted − risk scoreuntransmitted]/risk scoreuntransmitted) and compared between different discovery samples using two-sided paired t tests. For further details of both approaches, see Supplementary Methods in the online data supplement.
Results
Combined Tourette’s Syndrome-OCD GWAS
The final combined Tourette’s syndrome-OCD data set consisted of 2,723 cases (1,310 with OCD, 834 with Tourette’s syndrome, 579 with OCD and Tourette’s syndrome/chronic tics), 5,667 controls, and 290 OCD trios. A total of 7,659,573 SNPs (439,840 genotyped and 7,219,733 imputed) were included in the meta-analysis. The genomic control λ showed no evidence of residual population stratification or systematic technical artifacts (λGC=1.030; see Figure S3 in the online data supplement).
Sixty-eight SNPs with p<1×10−5, representing 16 independent genomic regions, were identified, although none reached the genome-wide significance threshold of p<5×10−8 (Table 1, Figure 1; see also Table S1 in the online data supplement). The most significant association was found in rs4988462 on 3p11 (p=3.72×10−7, odds ratio=1.18). This SNP lies within an intron of POU1F1, although the entire 279-kb region of association in LD with rs4988462 contains 16 additional SNPs with p<1×10−5 and includes CHMP2B and POU1F1 as well as the microRNA MIR4795. Regional association and forest plots from the top five independent GWAS signals are provided in Figures S4–S8 in the data supplement. Eleven of the 68 SNPs were also identified in the original OCD GWAS with p<1×10−5; none of these SNPs were identified in the Tourette’s syndrome GWAS at p<1×10−5 (15, 16) (see Table S1 in the data supplement).
| Chr | SNP | A1/A2 | A1 FRQ | Odds Ratio | Combined GWAS p | Position (hg19) | Number of SNPs in LD | Genes | Tourette’s Syndrome GWAS p | OCD GWAS p |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | rs4988462 | T/C | 0.42 | 1.18 | 3.7×10–7 | 87,127,019–87,406,369 | 343 | MIR4795, CHMP2B, POU1F1 | 1.1×10–3 | 4.9×10–5 |
| 11 | rs4271390 | T/C | 0.25 | 1.2 | 1.1×10–6 | 119,514,810–119,537,683 | 18 | PVRL1 | 1.1×10–3 | 4.4×10–5 |
| 13 | rs11149058 | C/T | 0.22 | 0.82 | 1.4×10–6 | 77,515,486–77,992,185 | 135 | IRG1, CLN5, Mir_633, FBXL3, MYCBP2 | 1.6×10–5 | 3.1×10–3 |
| 3 | rs149183310 | T/A | 0.044 | 1.52 | 1.5×10–6 | 115,864,394–116,185,436 | 77 | LSAMP | 3.1×10–3 | 3.4×10–5 |
| 3 | rs73070160 | T/C | 0.14 | 1.24 | 3.3×10–6 | 34,819,681–35,152,858 | 152 | FECHP | 1.3×10–3 | 3.8×10–4 |
| 18 | rs12959570 | G/A | 0.23 | 1.2 | 3.9×10–6 | 54,247,051–54,522,865 | 174 | TXNL1, WDR7 | 3.9×10–2 | 8.7×10–7 |
| 9 | rs7848024 | A/G | 0.27 | 1.19 | 5.3×10–6 | 105,520,114–105,753,694 | 248 | CYLC2 | 1.2×10–3 | 1.2×10–4 |
| 13 | rs6563569 | C/T | 0.44 | 1.16 | 5.7×10–6 | 38,087,567–38,290,276 | 238 | POSTN, TRPC4 | 3.0×10–4 | 3.3×10–3 |
| 6 | rs859980 | T/C | 0.47 | 0.87 | 6.5×10–6 | 104,462,081–104,517,073 | 93 | LOC100129694 | 8.6×10–3 | 5.0×10–5 |
| 9 | rs10973956 | A/C | 0.16 | 1.23 | 7.2×10–6 | 38,643,129–38,677,136 | 29 | FAM201A | 5.6×10–4 | 1.0×10–3 |
| 2 | rs35881094 | G/T | 0.43 | 1.16 | 7.4×10–6 | 58,847,436–59,058,234 | 122 | FLJ30838 | 5.9×10–4 | 7.6×10–4 |
| 11 | rs11021169 | T/G | 0.08 | 0.77 | 7.8×10–6 | 95,194,927–95,253,183 | 16 | 5S_rRNA | 1.7×10–4 | 4.5×10–3 |
| 20 | rs55797066 | C/T | 0.06 | 1.33 | 8.1×10–6 | 50,165,634–50,442,187 | 25 | NFATC2, ATP9A, SALL4 | 7.2×10–4 | 5.4×10–5 |
| 1 | rs7524258 | T/C | 0.4 | 1.16 | 8.4×10–6 | 7,250,522–7,352,143 | 87 | CAMTA1 | 1.4×10–2 | 2.4×10–5 |
| 4 | rs61792199 | G/A | 0.15 | 1.23 | 8.6×10–6 | 22,833,517–22,935,712 | 53 | GBA3 | 5.4×10–4 | 1.1×10–3 |
| 14 | rs1040832 | A/G | 0.2 | 1.21 | 9.9×10–6 | 93,247,868–93,374,784 | 33 | GOLGA5, CHGA | 5.1×10–3 | 2.6×10–4 |
TABLE 1. Genomic Regions With p<1×10–5 in the Combined Tourette’s Syndrome-Obsessive-Compulsive Disorder (OCD) Genome-Wide Association Study (GWAS)a
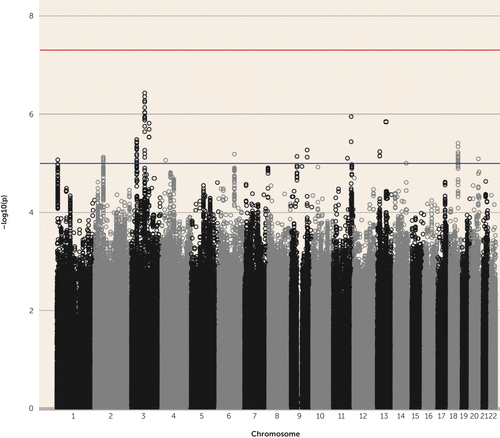
FIGURE 1. Manhattan Plot of All Genotyped and Imputed Single-Nucleotide Polymorphisms for 3,013 Tourette’s Syndrome-Obsessive-Compulsive Disorder European Ancestry Cases and 5,957 Controlsa
a Red and blue lines indicate significance thresholds of 5×10−8 and 1×10−5, respectively.
Enrichment Analyses
For Tourette’s syndrome, OCD, and the combined sample, we examined the subset of disease association p values for SNPs meeting stringent criteria for eQTLs (peQTL <10−6) derived from cerebellum, parietal cortex, and frontal cortex, as well as cerebellar mQTLs (pmQTL <10−6) (Figure 2). Using the field standard false-discovery-rate threshold of <0.25, we identified 38 cerebellar eQTLs from five LD-independent loci for Tourette’s syndrome, 161 cerebellar mQTLs (19 LD-independent loci) for OCD, and 53 parietal cortex eQTLs (four LD-independent loci) for the combined GWAS (Table 2).
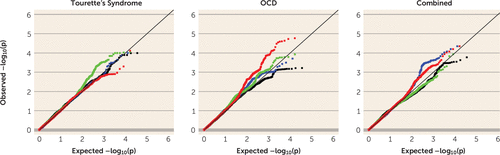
FIGURE 2. Q-Q Plot of Nominal Disease Association p Values Versus Expected p Values Among the Cis eQTLs and mQTLs in Different Brain Tissues in the Tourette’s Syndrome, Obsessive-Compulsive Disorder (OCD), and Combined Genome-Wide Association Study (GWAS)a
a eQTL=expression quantitative trait locus; mQTL=methylation quantitative trait locus. A horizontal shift to the left from the diagonal line (of complete concordance between the observed p values and expected p values under the null hypothesis of no enrichment) in the Q-Q plot indicates enrichment. Red dots represent cerebellum mQTLs, green dots represent cerebellum eQTLs, black dots represent frontal cortex eQTLs, and blue dots represent parietal cortex eQTLs.
| Tourette’s Syndrome GWAS | OCD GWAS | Combined GWAS | |||||
|---|---|---|---|---|---|---|---|
| Tissue | Functional Subset | QTLs | Loci | QTLs | Loci | QTLs | Loci |
| Frontal cortex | eQTLs | 0 | 0 | 0 | 0 | 0 | 0 |
| Parietal cortex | eQTLs | 0 | 0 | 0 | 0 | 53 | 4 |
| Cerebellum | eQTLs | 38 | 5 | 0 | 0 | 0 | 0 |
| Cerebellum | mQTLs | 0 | 0 | 161 | 18 | 0 | 0 |
TABLE 2. Number of Associated eQTLs/mQTLs With False Discovery Rate <0.25 in the Tourette’s Syndrome, Obsessive-Compulsive Disorder (OCD), and Combined Genome-Wide Association Study (GWAS)a
Polygenic Risk Score Analysis
Polygenic score analyses were conducted to test two related hypotheses: 1) that both Tourette’s syndrome and OCD individually harbor multiple, small-effect, common risk alleles across the genome; and 2) that Tourette’s syndrome and OCD may have shared common risk alleles (cross-disorder analyses). In the individual disorder analyses, risk scores derived from the OCD without known co-occurring Tourette's syndrome/chronic tics discovery sample strongly predicted case-control status in the OCD target sample (p=2.1×10−4), explaining 3.2% of the phenotypic variance (Figure 3; see also Table S3 in the online data supplement). In contrast, risk scores derived from the Tourette’s syndrome discovery sample demonstrated only weak prediction in the Tourette’s syndrome target sample (p=0.06; R2=0.6% of variance explained). Risk scores derived from the combined Tourette’s syndrome-OCD discovery sample also predicted case-control status in the OCD target sample (p=0.0075, R2=1.7% of variance explained), although less robustly than those derived from the OCD discovery sample alone (p=0.01; see Figure 3, inset). Risk scores derived from the Tourette’s syndrome-OCD combined sample could not discriminate between cases and controls in the Tourette’s syndrome target sample (p=0.4; see Figure 3; see also Table S3 in the data supplement).
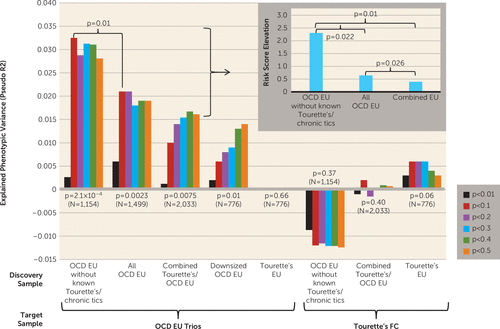
FIGURE 3. Individual Disorder and Cross-Disorder Polygenic Score Analysis in Tourette’s Syndrome and Obsessive-Compulsive Disorder (OCD)a
a The variance explained in two target samples (OCD European ancestry [EU] parent-child trios and Tourette’s French Canadian [FC] cases and matching controls) is based on risk scores derived from an aggregated sum of weighted single-nucleotide polymorphism risk allele effect sizes estimated from discovery samples at six significance thresholds. The y axis indicates Nagelkerke’s pseudo R2. The p value under each discovery sample indicates how well the risk scores derived from the discovery sample can predict the illness phenotype in the target sample. N is the number of cases in each discovery sample. Negative R2 values indicate a negative correlation between risk scores and illness status in the target sample. OCD EU without known Tourette’s/chronic tics=European-ancestry OCD genome-wide association study (GWAS) samples after removing samples with known co-occurring Tourette’s/chronic tics; all OCD EU=European-ancestry OCD GWAS samples plus additional EU GWAS samples with co-occurring OCD and Tourette’s/chronic tics; combined Tourette’s/OCD EU=all European-ancestry Tourette’s GWAS samples and OCD GWAS samples; downsized OCD EU=randomly selected subset of OCD EU samples to match the number of cases in the Tourette’s EU discovery sample; Tourette’s EU=European-ancestry Tourette’s GWAS samples; OCD EU trios=the OCD EU parent-child trio probands and matched pseudo-control data derived from nontransmitted alleles; Tourette’s FC=Tourette’s French Canadian cases and matching controls. A permutation test was carried out to determine the significance of the difference in R2 between risk scores derived from OCD EU without known Tourette’s/chronic tics and all OCD EU, resulting in a two-sided empirical p value of 0.01. The inset box at upper right demonstrates the risk score elevations (difference in risk scores of transmitted alleles and untransmitted alleles in the OCD EU trios, standardized by the risk score of the untransmitted alleles) derived from three discovery samples: OCD EU without known Tourette’s/chronic tics, all OCD EU, and combined Tourette’s/OCD EU. Two-sided paired t tests were conducted for the pairwise comparisons of risk score elevations derived from three discovery samples.
In cross-disorder analyses, risk scores derived from the Tourette’s syndrome discovery sample did not predict case-control status in the OCD target sample (p=0.66), nor did OCD-associated risk scores predict into the Tourette’s syndrome target sample (p=0.37) (see Figure 3 and Table S3).
To explore the influence of phenotype comorbidity on polygenic risk score prediction, an additional all-OCD discovery sample was created that included the primary OCD discovery sample plus 345 additional case subjects with OCD plus co-occurring Tourette’s syndrome/chronic tics. As expected, the polygenic score using risk alleles derived from this discovery sample predicted case-control status in the OCD target sample (p=2.3×10−3) (Figure 3). However, the proportion of variance explained by the all-OCD risk score was significantly attenuated compared with the risk score derived from the primary OCD without co-occurring Tourette's syndrome/chronic tics discovery sample, despite the 30% increase in sample size (OCD without co-occurring Tourette's syndrome/chronic tics, N=1,154, R2=3.2% of variance explained; all-OCD sample, N=1,499, R2=2.1% of variance explained; permutation p=0.01; see Figure 3; see also Figure S9 in the data supplement).
In addition, the magnitude of elevation in the polygenic risk scores (risk score elevation) between transmitted and untransmitted risk alleles in the OCD trios was calculated using risk alleles from the different OCD discovery samples and compared (see Figure 3, inset). The risk score elevation in the OCD trios was highest when the primary OCD without co-occurring Tourette's syndrome/chronic tics discovery sample was used to derive the risk score compared to either the all-OCD sample or the combined Tourette’s syndrome-OCD sample (paired t test, p=0.022 and p=0.010, respectively), consistent with a dilution of risk when either OCD cases with Tourette’s syndrome/chronic tics or Tourette’s syndrome cases without OCD were incorporated in the discovery sample.
Discussion
Our goal in this study was to leverage phenotypic and genotypic data of two phenotypically related and frequently co-occurring neurodevelopmental disorders, Tourette’s syndrome and OCD, to explore the hypothesis that these disorders share common genetic susceptibility variants. Our strategy was 1) to combine the samples in a joint GWAS, 2) to examine their patterns of eQTL/mQTL enrichment, and 3) to explore cross-disorder polygenic signals. Although limited by small sample sizes, the results of these diverse analytic approaches suggest a complex genetic relationship between Tourette’s syndrome and OCD.
While our previous work with this sample provides evidence of genetic sharing between Tourette’s syndrome and OCD, with a genetic correlation of 0.41 between the two disorders (10), we did not identify any genome-wide significant variants for the combined Tourette’s syndrome-OCD phenotype in this GWAS analysis, despite the increase in sample size. However, the combined GWAS signals were significantly enriched for functional alleles (parietal eQTLs), suggesting that these subthreshold variants contain some proportion of Tourette’s syndrome-OCD risk loci that are not simply due to stochastic variation. In the presence of genetic heterogeneity (see below), this sample is underpowered to determine whether these loci contribute to susceptibility to both Tourette’s syndrome and OCD, or to susceptibility to one or the other individually. As with any genetic association result, replication in an independent sample is required to know whether any of the individual eQTLs identified here are truly shared Tourette’s syndrome-OCD susceptibility variants (9, 26, 27).
However, the results of the polygenic analyses do provide strong evidence that OCD and Tourette’s syndrome have at least some distinct genetic risk factors. First, the individual disorder analyses confirm that OCD has a significant polygenic component. The proportion of OCD variance explained by directly interrogated SNPs (3.2%) is similar to the findings in schizophrenia (3%–6%) and bipolar disorder (2.8%) (25), indicating that OCD likely arises from the joint influence of a large number of susceptibility genes spread across the genome, either as common variants or as rare variants in tight linkage disequilibrium with GWAS SNPs. This result is consistent with a parallel heritability study of the same data sets using mixed linear modeling, which found that OCD heritability is concentrated in common variants with minor allele frequencies >30% (10).
In contrast, the proportion of Tourette’s syndrome variance explained was substantially lower (0.6%). Although some of the difference in polygenic risk prediction between OCD and Tourette’s syndrome may be due to the smaller discovery sample size for Tourette’s syndrome, a sensitivity analysis in which the OCD discovery sample size was reduced to match that of the Tourette’s syndrome sample still detected a larger, and statistically significant, OCD polygenic signal than the comparable Tourette’s syndrome signal (p=0.01) (see Figure 3 and Table S3). The Tourette’s syndrome discovery sample was also too small to examine polygenic signals in Tourette’s syndrome subgroups (Tourette’s syndrome plus OCD versus Tourette's syndrome without OCD); thus, it is possible that the Tourette’s syndrome polygenic signal could increase if Tourette’s-syndrome-only discovery and target samples were available. The Tourette’s syndrome polygenic signal may also have been attenuated by restricting polygenic risk score SNPs to those with minor allele frequencies >5% (done to reduce bias due to undercalling of rare variants; see Supplementary Methods in the data supplement), as this class of SNPs has been shown to account for ∼20% of the variance in liability to Tourette’s syndrome, with 80% attributable to common variants (10). Both the investigation of Tourette’s syndrome subgroups and the analysis of polygenic signal including SNPs with minor allele frequencies ≤5% may be possible in the future as the number of subjects with available GWAS data increases.
The cross-disorder polygenic analyses also provide evidence for genetic heterogeneity between OCD and Tourette’s syndrome. First, the polygenic risk scores generated from the individual OCD and Tourette’s syndrome discovery samples did not predict case-control status of the other disorder. Second, the combined Tourette’s syndrome-OCD sample was a worse predictor of OCD or Tourette’s syndrome status than either disorder alone, suggesting that the degree of genetic heterogeneity generated by combining the two phenotypes outweighs any improvement in statistical power due to increased sample size. As noted above, however, our data are likely underpowered to detect a modest shared signal, which we have previously identified in this sample using a mixed-model approach (10).
Although we were not able to examine Tourette’s syndrome subgroups, we were able to examine the polygenic composition within OCD subgroups (OCD with or without Tourette’s syndrome/chronic tics). These results clearly suggest that OCD with and without chronic tics have different genetic architectures. When OCD cases with co-occurring Tourette’s syndrome/chronic tics were added to the OCD discovery sample, the polygenic signal in the independent OCD target sample was attenuated by 35% (permutation p=0.01), despite the 30% increase in sample size. Similarly, the risk score elevation between transmitted and untransmitted alleles dropped substantially with the addition of these 345 OCD cases with co-occurring Tourette’s syndrome/chronic tics (p=0.022).
The hypothesis that OCD may be genetically heterogeneous, with some individuals and families segregating OCD without tics and others a subtype of OCD with tics that may share genetic risk with Tourette’s syndrome, was originally proposed by Pauls et al. in 1986 (27), and more recent epidemiologic studies have provided additional support for this concept (9, 26, 28). Although not yet studied, these genetic differences may also correlate with well-documented differences in treatment outcomes of patients who have OCD alone compared with those who have OCD with tics, in which the latter are more refractory to treatment and may require augmentation with antipsychotics (29–31).
Limitations
The primary limitation of this study is related to sample size. While our study represents the largest genetic sample of either disorder studied to date, the total sample of 3,013 case subjects and 5,957 control subjects has 67% power to detect an illness variant with an odds ratio of 1.25 (assuming the risk allele frequency is 20% in the general population), and only 25% power to detect a variant with an odds ratio of 1.20. Recent studies of other psychiatric disorders with evidence of genetic overlap have required substantially larger sample sizes in order to detect individual variants that contribute to both disorders (32, 33). Therefore, caution is necessary when drawing conclusions about the genetic architecture of Tourette’s syndrome and OCD based exclusively on the results of the combined GWAS. However, we have more confidence in our interpretation of the polygenic analyses, which demonstrated significant differences between the aggregate polygenic risk for the Tourette’s syndrome-OCD phenotypes despite comparatively small sample sizes. Of note, aggregate polygenic signals have been successfully detected with a comparable number of subjects in other cross-disorder studies as well (32, 33).
In addition, although we propose that the differences in polygenic risk prediction between Tourette’s syndrome and OCD and between OCD with and without tics are due to divergent genetic architectures, alternative explanations should be considered, such as diagnostic misclassification or differences in case ascertainment between study sites or over time. It is also important to note that we focused on common variation, and that rare inherited variation, unique mutations within individual families, de novo mutations, structural variation, and epigenetic and nongenetic factors are all likely contributors to the overall etiology of these related disorders. While our initial studies suggested that common variants account for most of the heritability of Tourette’s syndrome and OCD (10), it is still critically important to explore all of these potential contributors to disease in order to acquire a full understanding of their relative contributions to Tourette’s syndrome and OCD.
Finally, interpretation of the eQTL/mQTL analyses is limited by the fact that the tissues analyzed represent a convenience sample based on currently available data, and hence conclusions about tissue specificity should be reserved until larger eQTL data sets across the full range of brain regions and developmental time periods are available.
Overall, our results argue that, in addition to some shared genetic variants contributing to susceptibility to either Tourette’s syndrome or OCD, genetic variants likely exist that provide phenotypic specificity for each disorder. This observation contrasts with the hypothesis that genes contributing to neuropsychiatric disorders provide a “generalist” framework of neuronal connections from which nongenetic factors determine specific phenotypes, as has been proposed to explain the wide range of phenotypes observed in patients with similar large recurrent copy number variants across various regions of the genome (34, 35). Furthermore, the apparent difference between OCD with and without tics supports the importance of detailed phenotypic characterization to identify subtype-specific risk alleles in the future. Collection of additional samples through ongoing collaboration will be crucial to further elucidate the specific underlying susceptibility genes for Tourette’s syndrome and OCD, both shared and unique.
1 : Probing striato-thalamic function in obsessive-compulsive disorder and Tourette syndrome using neuroimaging methods. Adv Neurol 2001; 85:207–224Medline, Google Scholar
2 : The genetics of obsessive compulsive disorder and Gilles de la Tourette’s syndrome. Psychiatr Clin North Am 1992; 15:759–766Crossref, Medline, Google Scholar
3 : Tourette’s and comorbid syndromes: obsessive compulsive and attention deficit hyperactivity disorder: a common etiology? Clin Psychol Rev 1999; 19:531–552Crossref, Medline, Google Scholar
4 : A family study of early-onset obsessive-compulsive disorder. Am J Med Genet B Neuropsychiatr Genet 2005; 136B:92–97Crossref, Medline, Google Scholar
5 : Obsessive-compulsive disorder in Tourette’s syndrome. Adv Neurol 2005; 96:249–261Medline, Google Scholar
6 : Latent class analysis of Gilles de la Tourette syndrome using comorbidities: clinical and genetic implications. Biol Psychiatry 2008; 64:219–225Crossref, Medline, Google Scholar
7 : Sensory phenomena in obsessive-compulsive disorder and Tourette’s disorder. J Clin Psychiatry 2000; 61:150–156Crossref, Medline, Google Scholar
8 : The genetics of Tourette syndrome: a review. J Psychosom Res 2009; 67:533–545Crossref, Medline, Google Scholar
9 : Familiality of Tourette syndrome, obsessive-compulsive disorder, and attention-deficit/hyperactivity disorder: heritability analysis in a large sib-pair sample. J Am Acad Child Adolesc Psychiatry 2011; 50:46–54Crossref, Medline, Google Scholar
10 : Partitioning the heritability of Tourette syndrome and obsessive compulsive disorder reveals differences in genetic architecture. PLoS Genet 2013; 9:e1003864Crossref, Medline, Google Scholar
11 : The genetics of obsessive compulsive disorder: a review of the evidence. Am J Med Genet C Semin Med Genet 2008; 148C:133–139Crossref, Medline, Google Scholar
12 : l-Histidine decarboxylase and Tourette’s syndrome. N Engl J Med 2010; 362:1901–1908Crossref, Medline, Google Scholar
13 : Additional support for the association of SLITRK1 var321 and Tourette syndrome. Mol Psychiatry 2010; 15:447–450Crossref, Medline, Google Scholar
14 : Tourette syndrome is associated with recurrent exonic copy number variants. Neurology 2010; 74:1583–1590Crossref, Medline, Google Scholar
15 : Genome-wide association study of Tourette’s syndrome. Mol Psychiatry 2013; 18:721–728Crossref, Medline, Google Scholar
16 : Genome-wide association study of obsessive-compulsive disorder. Mol Psychiatry 2013; 18:788–798Crossref, Medline, Google Scholar
17 : PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81:559–575Crossref, Medline, Google Scholar
18 : Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38:904–909Crossref, Medline, Google Scholar
19 : A map of human genome variation from population-scale sequencing. Nature 2010; 467:1061–1073Crossref, Medline, Google Scholar
20 : A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5:e1000529Crossref, Medline, Google Scholar
21 : METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26:2190–2191Crossref, Medline, Google Scholar
22 : Estimation of significance thresholds for genomewide association scans. Genet Epidemiol 2008; 32:227–234Crossref, Medline, Google Scholar
23 : Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol 2008; 32:381–385Crossref, Medline, Google Scholar
24 : Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet 2010; 86:411–419Crossref, Medline, Google Scholar
25 : Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460:748–752Crossref, Medline, Google Scholar
26 : Complex segregation analysis for obsessive compulsive disorder and related disorders. Am J Med Genet 1999; 88:38–43Crossref, Medline, Google Scholar
27 : Gilles de la Tourette’s syndrome and obsessive-compulsive disorder: evidence supporting a genetic relationship. Arch Gen Psychiatry 1986; 43:1180–1182Crossref, Medline, Google Scholar
28 : Obsessive-compulsive disorder, tics, and anxiety in 6-year-old twins. Psychol Med 2007; 37:39–48Crossref, Medline, Google Scholar
29 : A systematic review: antipsychotic augmentation with treatment refractory obsessive-compulsive disorder. Mol Psychiatry 2006; 11:622–632Crossref, Medline, Google Scholar
30 : Tics moderate treatment outcome with sertraline but not cognitive-behavior therapy in pediatric obsessive-compulsive disorder. Biol Psychiatry 2007; 61:344–347Crossref, Medline, Google Scholar
31 : Tic-related vs non-tic-related obsessive compulsive disorder. Anxiety 1994–1995; 1:208–215Medline, Google Scholar
32 : Genome-wide association study identifies five new schizophrenia loci. Nat Genet 2011; 43:969–976Crossref, Medline, Google Scholar
33 : Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 2011; 43:977–983Crossref, Medline, Google Scholar
34 : Generalist genes: implications for the cognitive sciences. Trends Cogn Sci 2006; 10:198–203Crossref, Medline, Google Scholar
35 : Copy number variation in Tourette syndrome: another case of neurodevelopmental generalist genes? Neurology 2010; 74:1564–1565Crossref, Medline, Google Scholar



