A Genome-Wide Significant Linkage for Severe Depression on Chromosome 3: The Depression Network Study
Abstract
Objective:
The purpose of this study was to find loci for major depression via linkage analysis of a large sibling pair sample.
Method:
The authors conducted a genome-wide linkage analysis of 839 families consisting of 971 affected sibling pairs with severe recurrent major depression, comprising waves I and II of the Depression Network Study cohort. In addition to examining affected status, linkage analyses in the full data set were performed using diagnoses restricted by impairment severity, and association mapping of hits in a large case-control data set was attempted.
Results:
The authors identified genome-wide significant linkage to chromosome 3p25-26 when the diagnoses were restricted by severity, which was a maximum LOD score of 4.0 centered at the linkage marker D3S1515. The linkage signal identified was genome-wide significant after correction for the multiple phenotypes tested, although subsequent association mapping of the region in a genome-wide association study of a U.K. depression sample did not provide significant results.
Conclusions:
The authors report a genome-wide significant locus for depression that implicates genes that are highly plausible for involvement in the etiology of recurrent depression. Despite the fact that association mapping in the region was negative, the linkage finding was replicated by another group who found genome-wide-significant linkage for depression in the same region. This suggests that 3p25-26 is a new locus for severe recurrent depression. This represents the first report of a genome-wide significant locus for depression that also has an independent genome-wide significant replication.
Major affective disorders have a complex genetic etiology involving interplay of both genetic and environmental factors, including certain types of severe and threatening life events such as those associated with humiliation or loss (1). The heritability of depression is approximately 40% but increases to approximately 70% when twins with recurrent and severe major depression are examined (2, 3). Likewise, while the majority of studies suggest a relative risk (lambda) value of approximately 3 among siblings of patients with major depression (4), a study comparing the siblings of recurrent major depression patients with the siblings of healthy comparison subjects, using strict definitions of both depression and health, found a substantially higher relative risk (lambda: >9) (5). The only characteristics of probands associated consistently with higher familiality or heritability are recurrence of episodes and severity of disorder (2, 6).
To detect genetic loci that affect susceptibility to recurrent depression, the Depression Network (DeNt) study collected an initial sample of 417 families with 497 concordant sibling pairs to perform genome-wide linkage analysis (7). Using this first wave of families, suggestive and modest evidence for linkage was found on chromosomes 1p36, 12q, 13q, and 15q. The peaks on chromosomes 12q and 15q also showed evidence for linkage in other samples (8–10). In the present study, we report linkage analysis results using an expanded sample that includes 325 additional families contributing 474 sibling pairs, resulting in a total of 971 concordant sibling pairs with recurrent depression. The total sample also consisted of 118 discordant sibling pairs and 12 unaffected sibling pairs. Further details regarding the linkage analysis are presented in the data supplement accompanying the online version of this article.
Linkage can be easier to detect when heterogeneity and error in phenotype are decreased (11). For depression, reliability is improved by focusing on severe cases as indexed by symptom count and/or impairment (12). Furthermore, recurrence of episodes and severity of disorder are consistently associated with higher familiality or heritability (2, 6), and therefore we used the cumulative severity of the two most severe episodes to consider more severe depression in our linkage analysis. This linkage study was followed by a large case-control association study to identify involvement of single genes (13).
Method
Sibling-Pair Sample for Linkage Analysis
Sibling pairs affected with recurrent unipolar depression were recruited from eight clinical sites (Aarhus, Denmark; Bonn, Germany; Dublin; Lausanne, Switzerland; St. Louis; London; Cardiff, United Kingdom; and Birmingham, United Kingdom). Probands were all of European ancestry. Eligibility criteria were as follows: age ≥18 years; at least one affected sibling, not a monozygotic twin, also age ≥18 years; and both siblings experiencing ≥2 depressive episodes of at least moderate severity separated by at least 2 months of remission as defined by DSM-IV (14) or ICD-10-DCR (15) criteria. Pairs were excluded if either sibling had ever fulfilled criteria for mania, hypomania, or schizophrenia or if either experienced psychotic symptoms that were mood incongruent or present when there was no evidence of a mood disturbance. Other exclusion criteria were intravenous drug dependency and depression occurring solely in relation to alcohol use. All participants gave written informed consent for participation in the study. For further details regarding the study sample, see Farmer et al. (16).
Clinical Assessment
All participants were interviewed using the Schedules for Clinical Assessment in Neuropsychiatry (15, 17). Items of psychopathology in the interview were rated for presence and severity according to the worst and second worst episodes of depression identified by the participant. For rating severity, individuals were asked about the 4- to 6-week period when their symptoms were at their worst during each episode (peak intensity). The majority of items in the interview are coded as follows: 0=absence of the item; 1=present but to a mild degree or intermittently; 2=moderately severe and present for >50% of the peak intensity period or severe but present for <50% of the peak intensity period; and 3=severe for >50% of the peak intensity period. The computerized version of the interview (18) provides diagnoses according to DSM-IV and ICD-10-DCR. All interviewers from each site attended a 4-day training course, and all took part in a joint interrater reliability exercise. Further details regarding clinical assessment are provided by Farmer et al. (16).
Sibling-Pair Blood Samples for Linkage Analysis
At the time of interview, 25 ml of whole blood was collected in (ethylenediaminetetraacetic acid containing) Monovette tubes. In addition, drops of blood were placed on a Guthrie blood spot card. The blood samples were labeled with a bar code, gently mixed, and stored frozen upright in a –20°C freezer pending DNA extraction.
Genotyping was performed using standard methods by deCODE Genetics (Reykjavik, Iceland). Briefly, 1,130 microsatellite markers were typed in two waves of genotyping. A total of 350 individuals from wave 1 were regenotyped to ensure marker allele calling consistency. Seventy-five markers with Hardy-Weinberg equilibrium p values <0.001 were excluded. After exclusions, marker density was 3.3 cM, with a maximum gap of 10.2 cM. The average heterozygosity was 72.4%.
Sibling-Pair Linkage Analysis
All phenotypic information from interviews and questionnaires was coded and all blood samples were bar coded by assigning a number to each participant and removing any personal identifying information. The software program MERLIN (Multipoint Engine for Rapid Likelihood INference [19]) was used to perform nonparametric dichotomous linkage analysis. The deCODE, version 5, map was used for all analyses. Allele frequencies were estimated with the maximum likelihood option in MERLIN, using the entire sample. MERLIN multipoint LOD scores were calculated at each marker and at 10 positions between them. Specifically, we used MERLIN's algorithm for calculation of the Whittemore and Halpern nonparametric linkage pairs statistics (Z scores) (20), which the algorithm converts to LOD scores using the model described by Kong and Cox (21).
Linkage Analysis Descriptive
Relationships were examined using the graphical representation of relationships application (22), which detected half-siblings coded as full siblings (N=5), duplicate samples or monozygotic twins (N=3), and unexpected relatedness (N=14). PEDSTATS (23) was used to detect mendelization errors (N=674), which were found in 201 families (range: 1–18) across 303 markers (range: 1–54), with all genotypes (N=3,194) for the specific marker within the family showing the error recoded as fails. In addition, the “error” procedure was used to remove unlikely recombinants, while the “simulate” procedure was used to empirically estimate the false positive rate (1,000 simulations with the largest single family dropped to speed computation).
The general structure of the cohort consisted of sibling pairs without parents (58%) or sibling trios (16%). The average size of informative families was 2.45 persons, with the majority (82.1%) having one generation with two or more affected offspring. Families with three, four, and five or more affected offspring represented 17.7%, 5.4%, and 1.5% of the sample, respectively. Details of the number of probands and affected relatives genotyped from each collection site are presented in Table 1. Table 2 shows the clinical characteristics for each phenotype. Three different phenotypes were analyzed. In addition to affected status based on recurrent depression, two additional and more restrictive diagnostic categories were generated (severe recurrent depression and very severe recurrent depression). These categories were based on severity as measured by impairment. It should be noted that the minimum score for items on the Schedules for Clinical Assessment in Neuropsychiatry for each episode was 2, with a potential range of 0–3 (i.e., the sample was selected to have a certain minimum impairment severity for each episode). To generate the phenotypes, the impairment severity scores for the worst and second worst episodes from the assessment scale were summed. To be considered affected in the recurrent depression, severe recurrent depression, or very severe recurrent depression categories, participants needed to have summed severity scores of at least 4, 5, or 6, respectively, in addition to meeting diagnostic criteria for depression in each episode. Since the categories were based on a minimum score, the phenotypes were nested.
| Phenotype | ||||||
|---|---|---|---|---|---|---|
| Clinical Site | Number of Probands Ascertained | Age of Onset (Years) | Male (%) | Recurrent Depression | Severe Recurrent Depression | Very Severe Recurrent Depression |
| St. Louis | 732 | 19.1 | 24.9 | 575 | 262 | 95 |
| London | 233 | 21.1 | 24.5 | 222 | 170 | 84 |
| Dublin | 274 | 22.3 | 32.8 | 262 | 195 | 113 |
| Cardiff, United Kingdom | 248 | 24.9 | 25.8 | 232 | 162 | 96 |
| Birmingham, United Kingdom | 303 | 24.0 | 31.4 | 295 | 232 | 143 |
| Bonn, Germany | 276 | 29.8 | 25.4 | 260 | 249 | 238 |
| Lausanne, Switzerland | 208 | 24.6 | 27.9 | 194 | 126 | 46 |
| Aarhus, Denmark | 138 | 25.9 | 34.8 | 124 | 51 | 12 |
| Total | 2,412 | 2,164 | 1,447 | 827 | ||
TABLE 1. Demographic Characteristics and Genetic Phenotypes Among Affected Sibling Pairs With Recurrent Depression
| Phenotype | Total | Male (%) | Average Age of Onset (Years) |
|---|---|---|---|
| Recurrent depression | 2,164 | 26.12 | 22.6 |
| Severe recurrent depression | 1,447 | 26.14 | 23.1 |
| Very severe recurrent depression | 827 | 26.98 | 24.2 |
TABLE 2. Summary Statistics Across Three Different Genetic Phenotypes Among Affected Sibling Pairs With Recurrent Depression
Samples for Case-Control Analysis
Case patients and comparison subjects were drawn from two studies of recurrent depression (the Depression Case Control study and the Depression Network Study) using identical methods of case definition and the aforementioned phenotyping. The studies were approved by the local ethical committees, and written informed consent was obtained from all participants. The Depression Case Control sample consists of 1,346 recurrent depression patients (women: 69.3%) fulfilling DSM-IV and/or ICD-10 criteria of at least moderate severity and who were ascertained from the three clinical centers in the United Kingdom (London, Cardiff, and Birmingham). The mean age at onset was 22.9 years (SD=11.3; range: 1–62). Probands from the Depression Network Study were also analyzed (ascertained from all eight clinical sites). For a detailed description of these two samples, see the genome-wide association study (GWAS) conducted by Lewis et al. (24).
Comparison subjects (N=1,288) were contacted via the Medical Research Council general practice research framework and screened using a composite index of depressive and anxiety symptoms and with a telephone interview using the Past History Schedule. The proportion of women in this comparison group was 58.4%, and the mean age was 47.24 years (SD=12.2; range: 20–69). An additional 457 healthy volunteers (women, 61.4%), consisting of staff and students at King's College London, were also screened for mental health disorders using the Past History Schedule.
Genotyping for Case Patients and Comparison Subjects
Genotyping for case patients and comparison subjects was performed by the Centre National de Génotypage using the Illumina HumanHap610-Quad BeadChips (Illumina, Inc., San Diego), which contains 598,723 single nucleotide polymorphism (SNP) markers. Bar-coded DNA samples were received by the Centre National de Génotypage DNA banking facility in standard tubes, together with the sample information. All DNA samples were subjected to stringent quality control, and all processing was carried out under full laboratory information management system control.
Genotyping Workflow
The concentrations of all samples were adjusted to 50 ng/μl, and 15 μl of each sample was robotically dispensed into bar-coded 96-well plates. Samples from both case patients and comparison subjects were randomly distributed on the plates. The plates were processed in a fully automated Illumina BeadLab equipped with liquid-handling robots, Illumina BeadArray readers, and Illumina iScans. Genotyping was carried out using the Illumina Human610 quad array according to the manufacturer's recommendations. The raw data were analyzed using Illumina BeadStudio and extracted for statistical analysis.
Quality Control
Stringent quality control procedures were applied to individual and SNP data. Individuals were excluded if their genotypic data showed a missing rate >1%, abnormal heterozygosity, or a sex assignment that conflicted with phenotypic data or that they were related (up to second-degree relatives) to other study participants or were of non-European ancestry. SNPs with a minor allele frequency <1% or showing departure from Hardy-Weinberg equilibrium (p<1×10–5) were excluded. EIGENSTRAT analysis was performed again following quality control procedures, and the principal components showing significant differences in ancestry between case patients and comparison subjects were used as covariates in association testing (five components for case patients versus screened comparison subjects).
Statistical Analysis
Markers were selected for analysis if they were within the one-LOD interval of a genome-wide significant linkage. The primary test for association between SNPs and depression was logistic regression, including ancestry principal components, assuming a log-additive genetic model but also fitting a dominance-deviance model to allow for recessive or dominant effects. Analyses were carried out for all participants meeting the impairment severity restricted diagnosis (a summed Schedules for Clinical Assessment in Neuropsychiatry item score ≥4), with case patients not meeting these criteria being redefined as comparison subjects. Analysis was carried out using PLINK, version 1.07 (25).
Results
Genome-Wide Linkage for Depression Phenotypes
The genome scan results for all three nested phenotypes are shown in Figure 1 and were generated using all available informative families. The most significant results were observed on chromosome 3 for the impairment restricted diagnostic categories, with maximum LOD scores of 4.01 (for severe recurrent and very severe recurrent depression, respectively). The highest LOD score for recurrent depression was on chromosome 11 (LOD=1.79), and the highest LOD score for very severe recurrent depression was on chromosome 7 (LOD=1.91). One peak was genome-wide significant after correction for multiple testing at both the phenotype and marker level, with two adjacent markers achieving an empirical p value <0.05 after simulations. From the simulations, the 5% significance threshold was a LOD score of 3.53, and the suggestive threshold was a LOD score of 2.23, after accounting for the three phenotypes and 1,065 markers examined.
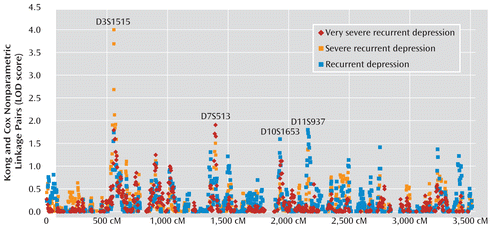
FIGURE 1. Autosome LOD Scores Among Affected Sibling Pairs in the Depression Network Study (Waves 1 and 2)
Chromosome 3
The most significant result observed in our combined analysis was on chromosome 3 using the severe recurrent depression diagnosis, which had a LOD of 4.0 at 19.9 cM (6.4 Mb [National Center for Biotechnology Information Build 36.1]) at marker D3S1515. In this analysis, affected was defined as depression with a minimum total impairment severity score of 5 (Figure 2). A LOD score of 4.0 corresponded with an empirical genome-wide p value of 0.015 after accounting for multiple testing at both markers and phenotypes via simulation of linkage (the proportion of times the observed LOD was exceeded by the maximum LOD scores from 1,000 simulations using MERLIN). Overall, the strongest marker was D3S1515 at 19.9 cM, with a LOD score of 4.0 and an empirical p value of 0.015, while its proximal flanking marker (D3S3591) at 23.9 cM had a LOD score of 3.567 and an empirical p value of 0.038. The distal flanking marker of D3S1515 was D3S2397, with a LOD score of 3.093 and an empirical p value of 0.254, which was the third most significant result. There is only modest support for linkage and reduced allele sharing in the same region for the recurrent depression phenotype, with a LOD score of 1.54, as well as the very severe recurrent depression phenotype, with a LOD score of 1.06. The one-LOD region extends 17.4 cM between markers D3S3706 (multipoint LOD: 2.674) and D3S1263 (multipoint LOD: 2.844) and covers approximately 7.3 Mb on hg18.
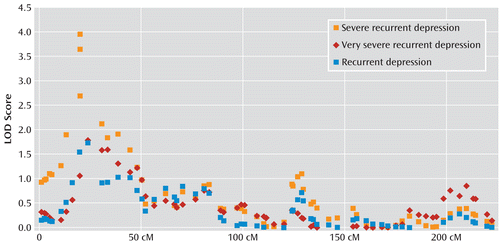
FIGURE 2. Chromosome 3 Linkage in the Depression Network Study (Waves 1 and 2)
In examining allele sharing in the sample at the maximum LOD marker (D3S1515) using the standardized Kong and Cox (21) allele sharing parameter delta, which MERLIN outputs, the model demonstrated that allele sharing increased when restricting to severe recurrent depression and remained the same when restricting further. Specifically, the Kong and Cox delta was 0.125 for recurrent depression, 0.229 for severe recurrent depression, and 0.234 for very severe recurrent depression. We note that while the allele sharing was similar between the two restricted diagnoses, the severe recurrent depression diagnosis had a much larger number of concordant sibling pairs and thus greater power.
Chromosome 7
No other peaks approached significance. The next most significant result after that observed on chromosome 3 was for very severe recurrent depression on chromosome 7, with a LOD score of 1.91 at 23.2 cM at marker D7S513 (11.6 Mb on hg18). This was the narrowest criteria used, and affected status was restricted to individuals with a maximum severity score for both their worst and second worst episodes. When taking into account the other two dichotomous phenotypes, the empirical genome-wide p value was 0.77. There was little support for linkage in the same region for the other diagnostic definitions, with an empirical p value of 1.0 for both phenotypes at this marker.
Other Regions
Selected regions such as chromosomes 1p, 12q, 13q, and 15q, which showed evidence for linkage in wave 1, were examined in detail for the recurrent depression phenotype (7). For chromosome 1p36, wave 1 showed a maximum multipoint LOD of approximately 2 at D1S450. However, there was no evidence for linkage in wave 2, and there was a maximum LOD of 0.75 in the combined wave 1 and 2 analysis (empirical p=1.0). Similar patterns of results were seen for chromosomes 12q and 13q, which showed no evidence for linkage in the combined sample. For chromosome 15, wave 1 had a maximum LOD of 2.44 at 91.8 cM (at marker D15S999), while the combined sample gave a maximum LOD of 1.41 (empirical p=0.98). Additional significant evidence for 15q has been reported after fine mapping with SNPs (26). However, this reported peak is approximately 21 cM distant.
Association Analyses
Genome-wide association data were available from 2,960 case patients and 1,594 screened comparison subjects (Figure 3). Logistic regression for SNP case-comparison association was performed in PLINK, version 1.06, adjusting for population stratification and dominance deviance for the severe recurrent depression phenotype, with a restriction to 5% minor allele frequency. After phenotype restriction, there were 1,590 case patients and 1,589 comparison subjects, meaning we compared intermediate severe case patients with screened comparison subjects and with other case patients being dropped from the analysis. There were 214 genes (including isoforms) in the University of California, Santa Cruz, genome browser annotation in the one-LOD interval between (hg18 coordinates) 3,895,145 and 11,636,572 on chromosome 3, as well as 1,878 SNPs, in our annotation. To allow for multiple testing and correlation between SNPs, empirical significance was calculated using the PLINK “mperm” procedure (100,000 case-comparison label permutations). Prior to permutation, no region-wide significant SNP or region-wide excess was observed, with 92 SNPs (4.89%/N=1,878) reaching a nominal level of significance (p<0.05). This remained the case after permutations, with the best SNPs being rs904467 and rs17050210, situated near SETD5, with an uncorrected p value <0.001 and lowest family-wise-corrected empirical p value of 0.19 for rs904467.
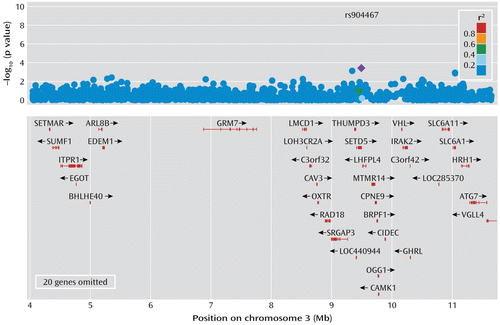
FIGURE 3. Association Analysis of the 3p25-26 Linkage Regiona
a The graph depicts chromosomal positions in mega base pairs (Mb). The association p value for each single nucleotide polymorphism (SNP) for severe recurrent depression is plotted. The purple diamond indicates the most significant SNP. (Because of space limitations, the following genes were omitted from the 8–11.5 Mb genes shown: TADA3, ARPC4, TTLL3, RPUSD3, JAGN1, IL17RE, IL17RC, CRELD1, PRRT3, TMEM111, LOC401052, CIDECP, FANCD2, C3orf24, C3orf10, TATDN2, GHRLOS, SEC13, ATP2B2, and MIR885.)
Discussion
We found a genome-wide significant linkage on chromosome 3p25-26, verified by simulations with a LOD score of 4.0 and an empirical p value of 0.015 at D3S1515 for severe recurrent depression. There is strong supporting evidence in that D3S1515's proximal flanking marker (D3S3591) also achieved genome-wide significance, and another flanking marker (D3S2397) had an empirical p value of 0.254 and was the third most significant result in the genome across the phenotypes tested.
There are multiple genes within the one-LOD region, including GRM7, which encodes metabotropic glutamate receptor mGluR7. Similar to its paralogues (mGluR4 and mGluR6), mGluR7 inhibits forskolin-stimulated cAMP accumulation in response to agonist interaction but is widely expressed in many neuronal cells of the CNS and plays an important role in modulation of glutamate transmission in the CNS (27). However, GRM7 is but one of the many strong candidate genes in this region. For example, ITPR1 is an inositol [1,4,5]-triphosphate receptor whose neuronal form is abundant in the cerebellum, particularly Purkinje cells (28), mutations which cause spinocerebellar ataxia type 15 (Mendelian Inheritance in Man number: 147265). Likewise, the oxytocin receptor gene OXTR is in the same region. Multiple reports have suggested lower levels of oxytocin in depression (29).
In order to resolve which gene or genes were important, we attempted fine mapping of this region in a large case-control cohort with identical phenotyping (including the Depression Network Study probands) by extracting genotypes within the one-LOD interval from available Illumina 610-quad genotyping (nearly 1,600 case patients with the same severe depression phenotype and just under 1,600 screened comparison subjects with lifetime and family absence of depression and other mental illness). No robust associations with either recurrent depression or the severity-restricted diagnoses were found in the region, suggesting that the linkage finding may be a result of multiple rare variants or that we may not have had sufficient power to detect multiple common variants of mild effect. Meta-analyses currently under way will have considerably greater power to detect effects in regions such as this.
The overall context of linkage and association findings in depression is not encouraging (30). We reviewed the literature on linkage studies, which included hits on chromosomes 1p, 12q, 13q, and 15q, and found that there has been little replication of depression linkage findings. A similar problem exists for depression candidate genes and GWASs, with large numbers of suggestive hits but no finding that has reached replicated genome-wide significance (24, 31).
The empirical p value of our chromosome 3 peak is 0.015, corresponding to a false positive rate of 1.5%. It is also a unique region in that it has not been reported in previous linkage studies for mood disorders. This and previous genetic findings in depression mean it is possible that our finding represents a false positive and requires replication and/or the identification of the causal variant(s) in the region. However, we can report that replication of our finding has been observed by Pergadia et al. (13), who report a genome-wide significant multipoint LOD score of 4.14 on chromosome 3 at 24.9 cM (3p25-3p26 [the same region as our finding]) in a sample of families of heavy smokers with depression.
The linkage peak we found in the Depression Network family sample and that found by Pergadia et al. (13) both exceed genome-wide significance. This appears to be one of the strongest replicated genetic findings in studies for depression. Follow-up work will include sequencing of the region in sibling pairs and in families sharing expanded haplotypes in the region as well as subsequent analyses of the region in additional samples with similar phenotyping.
1. : Humiliation, loss and other types of life events and difficulties: a comparison of depressed subjects, healthy controls and their siblings. Psychol Med 2003; 33:1169–1175Crossref, Medline, Google Scholar
2. : A hospital-based twin register of the heritability of DSM-IV unipolar depression. Arch Gen Psychiatry 1996; 53:129Crossref, Medline, Google Scholar
3. : A population-based twin study of major depression in women: the impact of varying definitions of illness. Arch Gen Psychiatry 1992; 49:257–266Crossref, Medline, Google Scholar
4. : Genetics of Affective Disorders. New York, Oxford University Press, 2002, pp 211–245Google Scholar
5. : Cardiff Depression Study: a sib-pair study of life events and familiality in major depression. Br J Psychiatry 2000; 176:150–155Crossref, Medline, Google Scholar
6. : Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry 2000; 157:1552–1562Link, Google Scholar
7. : Whole genome linkage scan of recurrent depressive disorder from the Depression Network Study. Hum Mol Genet 2005; 14:3337–3345Crossref, Medline, Google Scholar
8. : Genomewide significant linkage to recurrent, early-onset major depressive disorder on chromosome 15q. Am J Hum Genet 2004; 74:1154–1167Crossref, Medline, Google Scholar
9. : Novel loci for major depression identified by genome-wide association study of Sequenced Treatment Alternatives to Relieve Depression and meta-analysis of three studies. Mol Psychiatry 2011; 16:202–215Crossref, Medline, Google Scholar
10. : Predisposition locus for major depression at chromosome 12q22–12q23.2. Am J Hum Genet 2003; 73:1271–1281Crossref, Medline, Google Scholar
11. : Endophenotypes as quantitative risk factors for psychiatric disease: rationale and study design. Am J Med Genet 2001; 105:42–44Crossref, Medline, Google Scholar
12. : Stability of psychiatric diagnoses. an application to the affective disorders. Arch Gen Psychiatry 1992; 49:824–830Crossref, Medline, Google Scholar
13. : A 3p26-3p25 genetic linkage finding for DSM-IV major depression in heavy smoking families. Am J Psychiatry (published online May 16, 2011; doi:
14.
15.
16. : The Depression Network (DeNT) Study: methodology and sociodemographic characteristics of the first 470 affected sibling pairs from a large multi-site linkage genetic study. BMC Psychiatry 2004; 4:42Crossref, Medline, Google Scholar
17. : SCAN: Schedules for Clinical Assessment in Neuropsychiatry. Arch Gen Psychiatry 1990; 47:589–593Crossref, Medline, Google Scholar
18. : Computer Assisted Personal Interviewing Application for the Schedules for Clinical Assessment in Neuropsychiatry, Version 2.1, and Diagnostic Algorithms for World Health Organization ICD-10 Chapter V DCR and for Statistical Manual IV. Geneva, Switzerland, World Health Organization, 2003Google Scholar
19. : MERLIN: rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 2002; 30:97–101Crossref, Medline, Google Scholar
20. : A class of tests for linkage using affected pedigree members. Biometrics 1994; 50:118–127Crossref, Medline, Google Scholar
21. : Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 1997; 61:1179–1188Crossref, Medline, Google Scholar
22. : GRR: graphical representation of relationship errors. Bioinformatics 2001; 17:742–743Crossref, Medline, Google Scholar
23. : PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics 2005; 21:3445–3447Crossref, Medline, Google Scholar
24. : Genome-wide association study of major recurrent depression in the UK population. Am J Psychiatry 2010; 167:949–957Link, Google Scholar
25. : PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81:559–575Crossref, Medline, Google Scholar
26. : Genetics of recurrent early-onset major depression (GenRED): significant linkage on chromosome 15q25-q26 after fine mapping with single nucleotide polymorphism markers. Am J Psychiatry 2007; 164:259–264Link, Google Scholar
27. : Molecular characterization of a new metabotropic glutamate receptor mGluR7 coupled to inhibitory cyclic AMP signal transduction. J Biol Chem 1994; 269:1231–1236Medline, Google Scholar
28. : Ataxia and epileptic seizures in mice lacking type 1 inositol 1,4,5-trisphosphate receptor. Nature 1996; 379:168–171Crossref, Medline, Google Scholar
29. : Plasma oxytocin levels and anxiety in patients with major depression. Psychoneuroendocrinology 2007; 32:407–410Crossref, Medline, Google Scholar
30. : Genome-wide association of major depression: description of samples for the GAIN Major Depressive Disorder Study: NTR and NESDA biobank projects. Eur J Hum Genet 2008; 16:335–342Crossref, Medline, Google Scholar
31. : Poor replication of candidate genes for major depressive disorder using genome-wide association data. Mol Psychiatry (Epub ahead of print, March 30, 2010)Medline, Google Scholar



