Pharmacogenetics of Antidepressant Medication Intolerance
Abstract
OBJECTIVE: The authors sought to identify genetic markers for antidepressant medication intolerance. Genetic variation in drug metabolizing enzymes such as cytochrome P450 2D6 (CYP2D6) has been postulated to underlie antidepressant intolerance (pharmacokinetic effect). However, variation in genes encoding serotonin receptors could also explain antidepressant side effects (pharmacodynamic effect). METHOD: An 8-week, double-blind, randomized pharmacogenetic study compared the widely prescribed antidepressants paroxetine (a selective serotonin reuptake inhibitor [SSRI]) and mirtazapine (not an SSRI) in 246 elderly patients with major depression. Genotypes were determined for the 102 T/C single nucleotide polymorphism (SNP) in the serotonin 2A (5-HT2A) locus (HTR2A), previously associated with psychotropic medication treatment outcome. Oligonucleotide microarrays were used to extensively characterize variation in the CYP2D6 gene. Clinical outcomes included treatment discontinuations, adverse events, medication compliance, and change in mood. RESULTS: Survival analysis showed discontinuations due to paroxetine-induced side effects were strongly associated with the HTR2A C/C genotype. There was a significant linear relationship between the number of C alleles and the probability of discontinuation. Side effect severity in paroxetine-treated patients with the C/C genotype was also greater. In contrast, HTR2A 102 T/C genotype had no effect on mirtazapine side effects. CYP2D6 genotype did not predict treatment outcome for either medication. CONCLUSIONS: Pharmacodynamic differences among patients due to variant 5-HT2A receptors appear to be more important than pharmacokinetic variation in determining paroxetine intolerance. Pharmacogenetic markers may be useful in predicting antidepressant treatment outcome.
It is unknown why some patients experience side effects with antidepressant medications whereas others do not. Genetic differences among patients may contribute to medication intolerance. Variation in the CYP2D6 gene encoding debrisoquine hydroxylase can result in impaired antidepressant hepatic metabolism, leading to higher plasma concentrations and more side effects (1). However, genetically determined differences in receptor proteins, membrane transporters, and signal transduction molecules could also have important pharmacodynamic effects on antidepressant tolerability (2). For example, enhanced medication binding to a receptor variant that results in autonomic activation could increase side effects.
Selective serotonin reuptake inhibitor (SSRI) antidepressants such as paroxetine are among the most widely prescribed of all medications (3). These agents increase the availability of serotonin (5-HT) in the synaptic cleft (4, 5). The 5-HT2A receptor is widely distributed postsynaptically in the CNS and in the periphery (6–8). Activation of central 5-HT2A receptors may affect sleep and arousal (9, 10) and sexual behavior (11), whereas stimulation of peripheral 5-HT2A receptors affects gut motility and vascular smooth muscle tone (7, 8). SSRI side effects such as insomnia, agitation, gastrointestinal distress, and sexual dysfunction could be due to activation of 5-HT2A receptors, although other receptors such as 5-HT3 may also be involved. In contrast to SSRI medications, mirtazapine induces the release of norepinephrine as well as serotonin (12). In addition, mirtazapine blocks serotonin receptors of the 2 and 3 subclasses. This may be why mirtazapine results in fewer complaints related to sleep and arousal, sexual function, and the gut than are typically experienced with SSRIs.
We examined the effects of genetic variation at the CYP2D6 and HTR2A loci on adverse events and discontinuations during an 8-week comparison of paroxetine and mirtazapine treatment of elderly patients with major depression. The CYP2D6 gene is highly polymorphic, with more than 40 known alleles. Homozygosity for null alleles results in no debrisoquine hydroxylase activity, whereas one null allele in combination with an intermediate metabolic allele results in reduced activity. Initial doses of paroxetine are metabolized largely by debrisoquine hydroxylase, so poor and intermediate metabolizers might experience more adverse events (13). Debrisoquine hydroxylase also accounts for 25%–45% of mirtazapine clearance (14). The HTR2A 102 T/C single nucleotide polymorphism (SNP) has been associated with antipsychotic medication efficacy and side effects in some studies (15–17). Association of the HTR2A 102 T/C SNP with outcome after treatment with clozapine, which modified serotonergic activity in the brain, indicated that this polymorphism might also affect outcome with paroxetine and mirtazapine.
Method
The study was an 8-week, double-blind, randomized trial comparing mirtazapine and paroxetine conducted at 18 outpatient clinics in the United States. Institutional review board approval was obtained at each site, and all patients provided written informed consent. All patients (paroxetine: N=124, mirtazapine: N=122) were 65 years of age or older and free of major medical problems for at least 3 months. At screening, all met DSM-IV criteria for major depression (single or recurrent), had Mini-Mental State Examination (MMSE) (18) scores above the 25th percentile for their age, and had a 17-item Hamilton Depression Rating Scale (19) score of at least 18. Patients were excluded for clinically significant laboratory abnormalities, unstable medical conditions, drug or alcohol abuse, psychosis, recent suicide attempt, psychiatric conditions other than major depression, or antidepressant treatment within 7 days of commencing the study. Other concurrent medications were continued throughout the trial. Initial treatment was either 15 mg of mirtazapine (one active capsule and one placebo capsule) or 20 mg of paroxetine (two 10-mg capsules) given each evening. On day 14, doses were increased to 30 mg of mirtazapine or paroxetine. At days 28 and 42, dose increases to 45 mg of mirtazapine or 40 mg of paroxetine were allowed if the patient had not achieved Clinical Global Impression (20) change scores indicating “much improved” or “very much improved.” Patients were evaluated after 1, 2, 3, 4, 6, and 8 weeks of treatment. A meeting was held for investigators from the different sites before the onset of the study for assessment training and standardization of techniques. Additional information on clinical aspects of the study has been published (21).
Genomic DNA was extracted from EDTA-treated whole blood (Puregene kit, Gentra Systems, Minneapolis). Sixteen CYP2D6 alleles were queried using oligonucleotide microarrays according to the manufacturer’s instructions (GeneChip CYP450, Affymetrix, Santa Clara, Calif.) as described (22). CYP2D6 deletions and duplications and the *41 allele were identified as described (22, 23). Predicted phenotypes were determined by using published allele classifications (24, 25). Patients with two null alleles were designated poor metabolizers. Those with one null allele and an intermediate metabolic allele or two intermediate metabolic alleles (*41, *10, *9) were designated intermediate metabolizers. Patients with a duplication of *1 or *2 were designated ultrametabolizers. All other patients were designated extensive metabolizers. Genotype for the 102 T/C DNA single nucleotide polymorphism in the HTR2A gene was determined by using the method of Du et al. (26). Electrophoretic gels were scored by two observers blind to clinical data.
Mirtazapine concentrations in plasma were analyzed by using a liquid chromatographic assay with fluorescence detection after extraction of plasma using N-hexane. Paroxetine was assayed in plasma samples using an ultraviolet high-performance liquid chromatography method.
Treatment discontinuations were classified as due to any event and due to an adverse event. Severity of adverse events were given ratings of 1 (mild), 2 (moderate), or 3 (severe). These scores were summed and were standardized for treatment duration and average daily dose. Actual medication taken was determined by counting the number of prescribed tablets remaining at each clinic visit. Dosing compliance was determined by total number of medication doses taken divided by total number of capsules given. For comparison of medication and genotype groups on baseline demographic measures, baseline mood scales, MMSE, plasma drug concentrations, final daily dose, dosing compliance, and severity of adverse events, a general linear model analysis was used with center, treatment, and genotype as factors. To compare the number of subjects discontinuing because of adverse events in relation to genotype and treatment, Cochran-Mantel-Haenszel analyses were used. Kaplan-Meier survival analyses were used to compare probability of discontinuation between genotype groups. Mood was rated with the Hamilton depression scale and the Geriatric Depression Scale (27). The effect of genotype on improvement in mood was determined by using a general linear model analysis with drug, genotype, center, and baseline included in the model. To test for an association between CYP2D6 genotype and HTR2A genotype, chi-square tests were performed.
Sixty-six patients also took medications that are debrisoquine hydroxylase substrates or inhibitors during the study. To determine if concurrent medications interacted with CYP2D6 genotype to affect severity of adverse events, analyses of variance were performed with three factors: medication, CYP2D6 genotype, and concurrent medication.
Results
Effect of CYP2D6 Genotype
We detected 11 CYP2D6 alleles and 33 genotypes in 241 patients using oligonucleotide microarrays and additional assays for the *41 and *5 alleles and gene duplications. CYP2D6 allele frequencies are given in Table 1. Frequencies of the common CYP2D6 alleles (*1, *2 and *41, *3, *4, *5) did not differ significantly (two-by-five chi-square test) from those reported for Caucasian populations (28). The *10B allele was overrepresented in our sample, most likely because there were 15 ethnic minority patients included in the sample genotyped for CYP2D6.
There were 42 patients (17.4%) with genotypes encoding poor (N=16) and intermediate metabolism (N=26). Ten patients (4.1%) carried gene duplications encoding ultrametabolism. There was no significant difference between the treatment groups in the frequencies of CYP2D6 genotype groups. Because of the small number of poor metabolizers and ultrametabolizers, analyses were performed on two combined groups: poor and intermediate metabolizers versus extensive metabolizers and ultrametabolizers. There were no significant differences between subjects classified as poor or intermediate metabolizers and subjects classified as extensive metabolizers or ultrametabolizers in age, gender distribution, ethnicity, baseline body weight, baseline MMSE, or baseline depression rating scale scores (Table 2). For both medications, patients with genotypes predicting poor or intermediate metabolism showed no differences in the severity of adverse events or the frequency of discontinuations from those with genotypes encoding extensive and ultrametabolism. There were no differences between groups in final daily dose achieved or in dosing compliance. Plasma drug levels obtained after 4 weeks of treatment showed no significant differences for either drug between patients with predicted poor or intermediate metabolism and others. CYP2D6 genotype had no effect on depression measures for either drug. Reanalysis of data with patients carrying one null allele and a functional allele (N=64) classified as intermediate metabolizers yielded identical results. Results were similar when data for the 222 Caucasian patients with CYP2D6 genotypes were analyzed alone.
An analysis of variance showed no significant interaction between concurrent medication (debrisoquine hydroxylase inhibitor or substrate) and CYP2D6 genotype effects on severity of adverse events. There was no interaction between concurrent medication and study medication and no significant three-way interaction. These results indicate that concurrent medication did not interact with CYP2D6 genotype or study medication to affect the severity of adverse events.
Effect of HTR2A Genotype
In the full sample, HTR2A 102 T/C allele frequencies were C=0.575, T=0.425. Genotype frequencies did not differ significantly from Hardy-Weinberg equilibrium. We analyzed clinical results by comparing patients with the C/C genotype with others because this dichotomy was shown to predict outcome in patients treated with clozapine (15). Of paroxetine-treated patients, 41 (33.6%) had the C/C genotype, whereas 81 (66.4%) were T/C or T/T. For mirtazapine, there were 38 patients (30.6%) with the C/C genotype, whereas 86 (69.4%) were T/C or T/T. These frequencies were not significantly different between the paroxetine and mirtazapine treatment groups. There were no significant differences between patients with the C/C genotype and others in age, gender distribution, ethnicity, baseline body weight, plasma drug concentrations, baseline cognition, or severity of depression at baseline for either treatment group (Table 3).
Unlike CYP2D6 variation, the HTR2A 102 T/C SNP had a major effect on paroxetine side effects and discontinuations. There were significantly more discontinuations due to adverse events for C/C paroxetine-treated patients (46.3%) than for those with the T/C and T/T genotypes (16%) (Cochran-Mantel-Haenszel test: χ2=12.8, df=1, p=0.001). Survival analyses showed that paroxetine-treated patients with the C/C genotype were significantly more likely to discontinue treatment because of adverse events than were other patients at all assessment points (p=0.001 for all points, log rank chi-square tests) (Figure 1). The severity of side effects in paroxetine-treated patients with the C/C genotype was also greater (F=4.61, df=1, 179, p=0.03). During the first week of treatment, C/C patients took significantly less paroxetine than others (F=7.64, df=1, 108, p=0.007), indicating these patients did not comply with medication instructions. Paroxetine-treated patients discontinued early because of gastrointestinal complaints (vomiting, nausea, diarrhea) (N=7), somnolence and difficulty concentrating (N=7), agitation and sleep disturbance (N=6), and other side effects (dizziness, sweating, headache, sexual dysfunction) (N=7).
In contrast, among mirtazapine-treated patients, there were no differences between C/C patients and other subjects in severity of adverse events, final daily dose, dosing compliance, plasma levels, early discontinuations, or dropouts due to adverse events. Survival analysis showed no difference between those with the C/C genotype and others in discontinuations at any point during the study (Figure 1).
We also performed three-level analyses comparing patients with C/C, T/C, and T/T genotypes. For the full sample, genotype frequencies were as follows: C/C=32.1%, T/C=50.8%, and T/T=17.1%. There was no significant difference between the treatment groups in the distribution of HTR2A genotypes (paroxetine group: C/C=33.6%, T/C=51.6%, T/T=14.8%; mirtazapine group: C/C=30.6%, T/C=50.0%, T/T=19.4%). There were no significant differences among the genotype groups in age, gender distribution, ethnicity, body weight, or baseline clinical measures for either treatment group. Survival analyses showed that among paroxetine-treated patients, there was a positive linear relationship between the number of C alleles and the probability of discontinuation because of adverse events at all assessment points (p=0.025–0.009, log rank chi-square tests). No effect of HTR2A genotype was seen among mirtazapine-treated patients.
Although there were no significant differences between HTR2A genotype groups in the frequency of ethnic minorities, to minimize error due to population stratification data for Caucasians (N=117 for mirtazapine, N=109 for paroxetine) were analyzed separately. There were no significant differences between C/C carriers and other Caucasian patients in baseline demographics and clinical characteristics for either treatment group. For paroxetine-treated patients, survival analyses showed that C/C patients had a significantly greater probability of discontinuation due to adverse events at every assessment point in comparison with other patients (p<0.005 for all points, log rank chi-square tests), whereas among mirtazapine-treated Caucasian patients there was no difference.
Chi-square tests showed no association between HTR2A 102 T/C genotype and CYP2D6 genotype for the full cohort (Table 4), as well as when the paroxetine and mirtazapine cohorts were considered separately. This means it is unlikely that effects observed for HTR2A C/C genotype were due to an association with a particular CYP2D6 genotype category.
Discussion
These results demonstrate a major effect of the HTR2A 102 T/C polymorphism on adverse events in elderly depressed, nondemented patients treated with paroxetine but not among those treated with mirtazapine. The HTR2A 102 T/C SNP does not result in an amino acid substitution in the receptor protein. This indicates the effect we found on intolerance to paroxetine is due to linkage disequilibrium with another nearby variant that alters receptor function. There are a number of other SNPs in the HTR2A gene, some of which are nonsynonymous (15). In our sample the HTR2A 102 T/C SNP was in complete linkage disequilibrium with the -1438 promoter polymorphism, which could affect receptor levels. A difference in the levels of the mRNAs encoding the HTR2A C and T variants was recently reported in the human brain (29). The linear relationship between the number of C alleles and discontinuations suggests a gene-dosage effect.
The 5-HT2A receptor is located on postsynaptic neurons and binds serotonin in the synaptic cleft. Anatomic and physiologic data suggest that some SSRI side effects such as sleep and circadian disturbances and sexual dysfunction could be mediated through 5-HT2A receptors (9–11). HTR2A variation may alter receptor numbers, affinity, or signal transduction. The 5-HT2A receptors are also found on smooth muscle cells in the gut and vasculature (7, 8). Carriers of the C/C genotype may have side effects including gastrointestinal upset and dizziness after paroxetine from altered activation of smooth muscle 5-HT2A receptors. Unlike paroxetine, mirtazapine blocks the 5-HT2A receptor, preventing interaction with serotonin. By blocking the receptor, mirtazapine could negate the effects of any 5-HT2A functional variation.
CYP2D6 variation is widely considered important in determining the severity of medication side effects. However, there are few studies to substantiate this belief. CYP2D6 genotype has strong effects on plasma levels of older tricyclic antidepressants (22). Because paroxetine is metabolized largely by debrisoquine hydroxylase (13), and because debrisoquine hydroxylase accounts for a substantial portion of mirtazapine metabolism (14), we expected patients with genotypes encoding impaired metabolism to show more severe adverse events and more discontinuations. Elderly patients are considered vulnerable to small increases in antidepressant levels. However, CYP2D6 genotype did not influence side effects from either paroxetine or mirtazapine, even taking into account the recently identified -1584 (-1496) C/G promoter variant encoding intermediate metabolic activity (24). The effects of the HTR2A 102 T/C polymorphism on paroxetine side effects and discontinuations cannot be attributed to CYP2D6 effects, since there was no association between HTR2A 102 T/C and CYP2D6 genotypes.
Contrary to recently published recommendations (30), we found no evidence that dosages of these medications should be adjusted for CYP2D6 poor and intermediate metabolizers. Further, 66 patients were taking debrisoquine hydroxylase inhibitors or substrates during the study. Concurrent therapy with paroxetine or mirtazapine did not affect discontinuations or adverse events, even in patients with intermediate metabolic genotypes expected to be vulnerable to treatment with medications interacting with debrisoquine hydroxylase. It is possible that with a larger sample or with different concurrent medications, a small but significant effect of CYP2D6 genotype on outcome might be detected. However, this effect is unlikely to approach the magnitude of that seen for HTR2A genetic variation.
These results indicate that variation in the HTR2A gene may be important in determining medication discontinuations in geriatric patients treated with paroxetine. If confirmed, it may be possible to identify patients who should avoid paroxetine or take reduced doses because of increased risk for discontinuation. CYP2D6 genetic variation does not appear to be a major factor in determining paroxetine or mirtazapine discontinuations and adverse events, indicating that pharmacodynamic effects such as receptor variation can be more important than pharmacokinetic factors.
 |
 |
 |
 |
Presented in part at the 40th annual meeting of the American College of Neuropsychopharmacology, Waikaloa Village, Hawaii, December, 9–13, 2001. Received Feb. 24, 2003; revision received June 16, 2003; accepted June 18, 2003. From the Neuroscience Research Laboratories, Department of Psychiatry and Behavioral Sciences, Stanford University School of Medicine; and Organon Pharmaceuticals, Inc., West Orange, N.J. Address reprint requests to Dr. Schatzberg, Department of Psychiatry and Behavioral Sciences, Stanford University School of Medicine, 401 Quarry Rd., Room 300, Stanford, CA 94305-5548; [email protected] (e-mail). Funding was provided by Organon Pharmaceuticals, Inc.; the National Association for Research on Schizophrenia and Depression; The Nancy Pritzker Network; and VA Medical Research (Sierra Pacific MIRECC). Personnel from the Organon Medical Services department played a major role in the design, conduct, and analysis of the trial, which was performed in accordance with guidelines for clinical trials of pharmaceutical agents established by U.S. regulatory authorities. The sponsor hired an independent contract research organization (Scirex, Inc., Horsham, Pa.) to implement the study, to oversee collection of blood samples, and to maintain the study database. Statistical analyses were designed and performed by Dr. Murphy and by employees of Scirex, Inc. All DNA work was performed in Dr. Murphy’s laboratory at Stanford University. The research contract permitted the investigators to publish the data without mandatory approval by the sponsor. The sponsor did review the manuscript and provided comments and suggestions. This work has resulted in a patent application that is pending. The authors thank Nina Pascoe and Jeremy Claassen for their technical assistance.
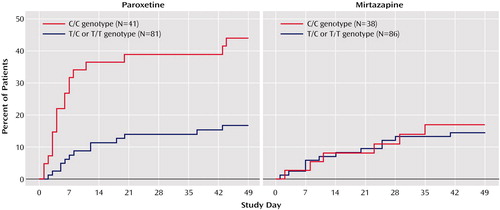
Figure 1. Treatment Discontinuation Among 246 Older Depressed Patients Randomly Assigned to 8 Weeks of Double-Blind Treatment With Paroxetine or Mirtazapine, by HTR2A Genotype for the 102 T/C Single Nucleotide Polymorphism
1. Bertilsson L, Dahl ML, Tybring G: Pharmacogenetics of antidepressants: clinical aspects. Acta Psychiatr Scand Suppl 1997; 391:14–21Crossref, Medline, Google Scholar
2. Cravchik A, Goldman D: Neurochemical individuality: genetic diversity among human dopamine and serotonin receptors and transporters. Arch Gen Psychiatry 2000; 57:1105–1114Crossref, Medline, Google Scholar
3. Access and Utilization of New Antidepressant and Antipsychotic Medications: Report to Office of the Assistant Secretary for Planning and Evaluation, US Department of Heath and Human Services. Washington, DC, US DHHS, Jan 2000Google Scholar
4. Bosker FJ, Klompmakers AA, Westenberg HG: Effects of single and repeated oral administration of fluvoxamine on extracellular serotonin in the median raphe nucleus and dorsal hippocampus of the rat. Neuropharmacology 1995; 34:501–508Crossref, Medline, Google Scholar
5. Malagie I, Deslandes A, Gardier AM: Effects of acute and chronic tianeptine administration on serotonin outflow in rats: comparison with paroxetine by using in vivo microdialysis. Eur J Pharmacol 2000; 403:55–65Crossref, Medline, Google Scholar
6. Fay R, Kubin L: Pontomedullary distribution of 5-HT2A receptor-like protein in the rat. J Comp Neurol 2000; 418:323–345Crossref, Medline, Google Scholar
7. Janssen P, Prins NH, Meulemans AL, Lefebvre RA: Pharmacological characterization of the 5-HT receptors mediating contraction and relaxation of canine isolated proximal stomach smooth muscle. Br J Pharmacol 2002; 136:321–329Crossref, Medline, Google Scholar
8. Banes A, Florian JA, Watts SW: Mechanisms of 5-hydroxytryptamine(2A) receptor activation of the mitogen-activated protein kinase pathway in vascular smooth muscle. J Pharmacol Exp Ther 1999; 291:1179–1187Medline, Google Scholar
9. Pullar IA, Carney SL, Colvin EM, Lucaites VL, Nelson DL, Wedley S: LY367265, an inhibitor of the 5-hydroxytryptamine transporter and 5-hydroxytryptamine(2A) receptor antagonist: a comparison with the antidepressant, nefazodone. Eur J Pharmacol 2000; 407:39–46Crossref, Medline, Google Scholar
10. Landolt HP, Meier V, Burgess HJ, Finelli LA, Cattelin F, Achermann P, Borbely AA: Serotonin-2 receptors and human sleep: effect of a selective antagonist on EEG power spectra. Neuropsychopharmacology 1999; 21:455–466Crossref, Medline, Google Scholar
11. Gorzalka BB, Hanson LA, Hong JJ: Ketanserin attenuates the behavioural effects of corticosterone: implications for 5-HT(2A) receptor regulation. Eur J Pharmacol 2001; 428:235–240Crossref, Medline, Google Scholar
12. de Boer T: The pharmacologic profile of mirtazapine. J Clin Psychiatry 1996; 57(suppl 4):19–25Google Scholar
13. Sindrup SH, Brosen K, Gram LF, Hallas J, Skjelbo E, Allen A, Allen GD, Cooper SM, Mellows G, Tasker TC, et al: The relationship between paroxetine and the sparteine oxidation polymorphism. Clin Pharmacol Ther 1992; 51:278–287Crossref, Medline, Google Scholar
14. Stormer E, von Moltke LL, Shader RI, Greenblatt DJ: Metabolism of the antidepressant mirtazapine in vitro: contribution of cytochromes P-450 1A2, 2D6, and 3A4. Drug Metab Dispos 2000; 28:1168–1175Medline, Google Scholar
15. Arranz MJ, Munro J, Birkett J, Bolonna A, Mancama D, Sodhi M, Lesch KP, Meyer JF, Sham P, Collier DA, Murray RM, Kerwin RW: Pharmacogenetic prediction of clozapine response. Lancet 2000; 355:1615–1616Crossref, Medline, Google Scholar
16. Joober R, Benkelfat C, Brisebois K, Toulouse A, Turecki G, Lal S, Bloom D, Labelle A, Lalonde P, Fortin D, Alda M, Palmour R, Rouleau GA: T102C polymorphism in the 5-HT2A gene and schizophrenia: relation to phenotype and drug response variability. J Psychiatry Neurosci 1999; 24:141–146Medline, Google Scholar
17. Segman RH, Heresco-Levy U, Finkel B, Goltser T, Shalem R, Schlafman M, Dorevitch A, Yakir A, Greenberg D, Lerner A, Lerer B: Association between the serotonin 2A receptor gene and tardive dyskinesia in chronic schizophrenia. Mol Psychiatry 2001; 6:225–229Crossref, Medline, Google Scholar
18. Folstein MF, Folstein SE, McHugh PR: “Mini-Mental State”: a practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 1975; 12:189–198Crossref, Medline, Google Scholar
19. Hamilton M: Development of a rating scale for primary depressive illness. Br J Soc Clin Psychol 1967; 6:278–296Crossref, Medline, Google Scholar
20. Guy W (ed): ECDEU Assessment Manual for Psychopharmacology: Publication ADM 76–338. Washington, DC, US Department of Health, Education, and Welfare, 1976, pp 218–222Google Scholar
21. Schatzberg AF, Kremer C, Rodrigues HE, Murphy GM Jr: Double-blind, randomized comparison of mirtazapine and paroxetine in elderly depressed patients. Am J Geriatr Psychiatry 2002; 10:541–550Crossref, Medline, Google Scholar
22. Murphy GM Jr, Pollock BG, Kirshner MA, Pascoe N, Cheuk W, Mulsant BH, Reynolds CF III: CYP2D6 genotyping with oligonucleotide microarrays and nortriptyline concentrations in geriatric depression. Neuropsychopharmacology 2001; 25:737–743Crossref, Medline, Google Scholar
23. Claassen JD, Pascoe N, Schatzberg AF, Murphy GM Jr: Rapid detection of the C-1496G polymorphism in the CYP2D6 *2 allele. Clin Chem 2001; 47:2153–2155Medline, Google Scholar
24. Raimundo S, Fischer J, Eichelbaum M, Griese EU, Schwab M, Zanger UM: Elucidation of the genetic basis of the common “‘intermediate metabolizer” phenotype for drug oxidation by CYP2D6. Pharmacogenetics 2000; 10:577–581Crossref, Medline, Google Scholar
25. Sachse C, Brockmoller J, Bauer S, Roots I: Cytochrome P450 2D6 variants in a Caucasian population: allele frequencies and phenotypic consequences. Am J Hum Genet 1997; 60:284–295Medline, Google Scholar
26. Du L, Bakish D, Lapierre YD, Ravindran AV, Hrdina PD: Association of polymorphism of serotonin 2A receptor gene with suicidal ideation in major depressive disorder. Am J Med Genet 2000; 96:56–60Crossref, Medline, Google Scholar
27. Yesavage JA, Brink TL, Rose TL, Lum O, Huang V, Adey M, Leirer VO: Development and validation of a geriatric depression screening scale: a preliminary report. J Psychiatr Res 1982–1983; 17:37–49Google Scholar
28. Marez D, Legrand M, Sabbagh N, Guidice JM, Spire C, Lafitte JJ, Meyer UA, Broly F: Polymorphism of the cytochrome P450 CYP2D6 gene in a European population: characterization of 48 mutations and 53 alleles, their frequencies and evolution. Pharmacogenetics 1997; 7:193–202Crossref, Medline, Google Scholar
29. Polesskaya OO, Sokolov BP: Differential expression of the “C” and “T” alleles of the 5-HT2A receptor gene in the temporal cortex of normal individuals and schizophrenics. J Neurosci Res 2002; 67:812–822Crossref, Medline, Google Scholar
30. Kirchheiner J, Brosen K, Dahl ML, Gram LF, Kasper S, Roots I, Sjoqvist F, Spina E, Brockmoller J: CYP2D6 and CYP2C19 genotype-based dose recommendations for antidepressants: a first step towards subpopulation-specific dosages. Acta Psychiatr Scand 2001; 104:173–192Crossref, Medline, Google Scholar



