Mood-Incongruent Psychotic Features in Bipolar Disorder: Familial Aggregation and Suggestive Linkage to 2p11-q14 and 13q21-33
Abstract
Objective: Mood-incongruent psychotic features in bipolar disorder may signify a more severe form of the illness and might represent phenotypic manifestations of susceptibility genes shared with schizophrenia. This study attempts to characterize clinical correlates, familial aggregation, and genetic linkage in subjects with these features. Method: Subjects were drawn from The National Institute of Mental Health (NIMH) Genetics Initiative Bipolar Disorder Collaborative cohort, consisting of 708 families recruited at 10 academic medical centers. Subjects with mood-incongruent and mood-congruent psychotic features were compared on clinical variables. Familial aggregation was tested using a proband-predictive model and generalized estimating equations. A genome-wide linkage scan incorporating a mood-incongruence covariate was performed. Results: Mood-incongruent psychotic features were associated with an increased rate of hospitalization and attempted suicide. A proband with mood-incongruence predicted mood-incongruence in relatives with bipolar I disorder when compared with all other subjects and when compared with subjects with mood-congruent psychosis. The presence of mood-incongruent psychotic features increased evidence for linkage on chromosomes 13q21-33 and 2p11-q14. These logarithm of the odds ratio (LOD) scores and their increase from baseline met empirical genome-wide suggestive criteria for significance. Conclusions: Mood-incongruent psychotic features showed evidence of a more severe course, familial aggregation, and suggestive linkage to two chromosomal regions previously implicated in major mental illness susceptibility. The 13q21-33 finding supports prior evidence of bipolar disorder/schizophrenia overlap in this region, while the 2p11-q14 finding is, to the authors’ knowledge, the first to suggest that this schizophrenia linkage region might also harbor a bipolar disorder susceptibility gene.
Mood-incongruent psychotic features in bipolar disorder may signify a more severe form of the illness with possible etiological ties to schizophrenia. The concept of mood-incongruence was derived by Karl Jaspers (1) , who differentiated between “delusions proper” and “delusion-like ideas,” depending on whether the content was “psychologically incomprehensible” or emerged “understandably from preceding affects.” Mood-incongruent psychotic symptoms in the presence of a mood or affective syndrome were initially classified under a broad schizophrenia diagnosis in the DSM-I and II, but were later categorized under the Research Diagnostic Criteria’s schizoaffective diagnosis. However, doubts about the diagnostic utility of mood-incongruence led to the reclassification of mood-incongruent psychotic mania or depression as a subset of affective disorder in the DSM-III, and this conception has been maintained in succeeding DSM editions.
Studies have found mood-incongruence to be common in mood disorder, with prevalence rates reported at 30%–40% of those with psychotic symptoms (2 , 3) . Although one study reported only modest interrater reliability for mood-incongruence (4) , more recent studies have reported kappa scores in the very good to excellent range (5 , 6) . Outcome investigations have found mood-incongruence to be associated with worse overall outcome (5 , 7) , poor lithium responsiveness (8) , and higher rates of attempted suicide (9) .
A few studies have provided modest evidence for familial aggregation between mood-incongruent psychotic mood disorder and schizophrenia. Two separate cohorts showed twice the morbid risk of schizophrenia in relatives of probands with mood-incongruent psychotic major depressive disorder compared with probands with the mood-congruent psychotic form of the illness, although these differences were not significant (10 , 11) . A third study reported insignificantly elevated rates of schizophrenia in relatives of probands with mood-incongruent psychotic major mood disorder relative to the family members of comparison subjects (12) . Conversely, in another report, relatives of probands with schizophrenia had almost five times the prevalence of mood-incongruent psychotic mood disorder compared with relatives of comparison subjects (13) .
If mood-incongruent psychotic symptoms have a genetic basis, they should cluster within a subset of bipolar disorder families. Although prior studies have shown evidence for familial aggregation of psychotic symptoms in bipolar disorder (14 , 15) , only one study, to our knowledge, has specifically addressed familial aggregation of mood-incongruent psychotic symptoms. In siblings with bipolar disorder, O’Mahony et al. (15) reported a modest correlation of mood-incongruent psychotic symptoms, which was statistically significant when all possible sibling pairs were evaluated but not when the analysis was restricted to independent pairs.
Molecular genetic investigations have suggested that bipolar disorder and schizophrenia may share some common susceptibility genes. Linkage studies have found signals in several overlapping chromosomal regions, including 6q16-22, 8p22-4, 13q31-34, 18p11, and 22q11-13 (16) . Association studies have also demonstrated overlap, particularly in the G72(DAOA)/G30 locus (17) and in the DISC1 gene (18) . A few studies have also provided evidence that the psychotic form of bipolar disorder in particular may be the phenotypic manifestation of overlap genes (19 , 20) . The only prior study to differentiate psychosis by mood-incongruence found that an apparent schizophrenia susceptibility haplotype in the neuregulin 1 gene was most strongly associated with bipolar disorder in subjects with more marked mood-incongruent psychotic symptoms (21) .
In this study, we hypothesized that the presence of mood-incongruent psychotic features would define a schizophrenia-like subset of bipolar disorder and that use of this trait would help identify overlap susceptibility genes. To test this hypothesis, we examined whether mood-incongruence would correlate with a more severe clinical course of bipolar disorder, aggregate in families and uncover genetic linkage in chromosomal regions previously associated with schizophrenia.
Method
Subject Ascertainment and Assessment
Bipolar disorder family cohorts were drawn from the National Institute of Mental Health (NIMH) Genetics Initiative Bipolar Disorder Collaborative project. The cohorts were ascertained in two phases between 1991 and 2003. The first phase was carried out at Johns Hopkins University, Baltimore; Indiana University, Bloomington, Indiana; Washington University in St. Louis; and the NIMH intramural program. The second phase was conducted by these four sites and additional sites at the University of California, San Diego; University of Iowa; University of Pennsylvania; University of Chicago; Rush-Presbyterian Medical Center, Chicago; and University of California, Irvine. The ascertainment criteria in both phases required a proband with bipolar I disorder and at least one sibling with either bipolar I disorder or schizoaffective disorder, bipolar type. Detailed ascertainment information has been published elsewhere (22) . Interviews were conducted using the Diagnostic Interview for Genetic Studies, versions 1.0, 2.0, or 3.0 (23) . Informed consent was obtained from all subjects after study procedures and goals were fully explained. Diagnoses were made with a best-estimate procedure using the DSM-III-R or DSM-IV and ranked on a 4-point confidence ordinal scale. Only diagnoses meeting confidence levels 3 (“confident”) or 4 (“high”) were included in the study. The clinical data collected in both phases were recently assembled into a single comprehensive database. This required that clinicians review items from the three versions of the Diagnostic Interview for Genetic Studies to determine where the versions differed and which questions could be combined across versions. Data managers reviewed responses to each item to identify inconsistencies, and clinicians reviewed potential errors, deeming data of uncertain quality as “unknown.” The cleaned dataset contained 3,643 subjects, including 708 probands, of whom 681 had bipolar I disorder (96.2%), and 27 had at least one sibling with either bipolar I disorder or schizoaffective disorder, bipolar type (3.8%).
Classification of Psychotic Features and Mood-Incongruence
The designation of psychotic features was based on a lifetime history of delusions or auditory or visual hallucinations. Subjects were only considered psychotic if their symptoms lasted persistently for at least 1 day or intermittently for 3 days or more. Mood-incongruence was defined according to DSM-IV as hallucinations or delusions with “content … inconsistent with depressive themes such as guilt, illness, personal inadequacy or catastrophe ... [or] inconsistent with manic themes such as inflated worth, power, knowledge, identity, or special relationship to a deity or famous person.” In the Diagnostic Interview for Genetic Studies, interviewers were asked to determine incongruence of content following descriptions of hallucinations or delusions in a mood episode. At least one positive response to these questions in either depressive or manic episodes rendered a subject positive for a lifetime history of mood-incongruence. Since DSM-IV considers “delusions of thought insertion … of thought broadcasting, and … of control” to be mood-incongruent, these psychotic symptoms, along with thought withdrawal, were also classified as mood-incongruent. Of note, interviewers deemed only a subset of persecutory delusions to be mood-incongruent.
Genotyping
Most of the ascertained families were genotyped genome-wide in four separate waves. Families in the first two waves were genotyped by the original four sites of the NIMH collaboration (24) . The last two waves of families were genotyped at the Center for Inherited Disease Research. The genotype data were cleaned using the following procedures: 1) the data were examined using the Pedigree Relationship Statistical Test to verify the reported familial relationships; 2) UNKNOWN and/or PedCheck were used to identify Mendelian inheritance errors; and 3) MERLIN was used to identify unlikely genotypes based on inferences about the gene flow in pedigrees given all available data. Potential errors were deleted if they could not be resolved. The cleaned genotype data from the four waves were combined into a single dataset. Markers were given a unique label for each wave of data and placed on a common genetic map using the deCode genetic map as a framework. Markers not available in the deCode map were placed on the framework according to their physical position in the July 2003 assembly of the Human Genome sequence. The genetic locations were then interpolated based on their physical position relative to the nearest flanking markers with known genetic locations. Identical markers genotyped in different waves were placed next to each other on the common map with the minimal distance between them. The resulting dataset used for the present linkage analyses included 2,899 genotyped subjects from 644 families, with a total of 669 markers, 366 of which were genotyped in more than one wave. These data have been analyzed for linkage with the standard bipolar disorder phenotype. (The report on this analysis by Zandi et al. is in preparation.) To facilitate comparison with prior findings, we report marker locations in this study based on the Marshfield map (http://research.marshfieldclinic.org/genetics/Map_Markers/maps/IndexMapFrames .html).
Data Analysis
Clinical correlates and familial aggregation
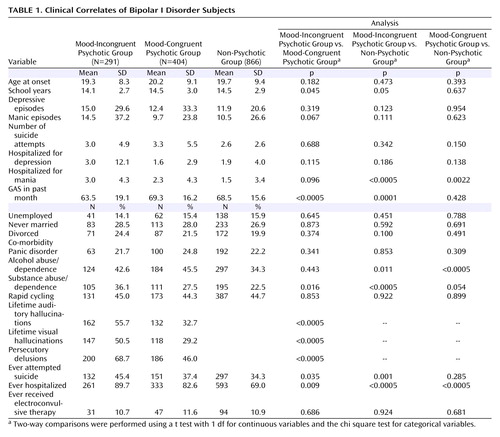
Differences between the three clinical subcategories ( Table 1 ) were analyzed with two-sided pairwise t tests for continuous data and Pearson’s chi square test for categorical data.
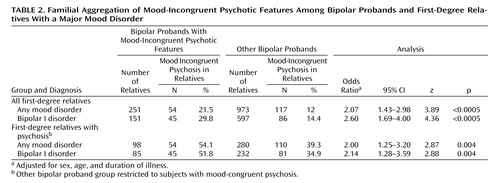
To assess familial aggregation, probands were initially divided into two groups: mood-incongruent psychotic features and nonmood-incongruent psychotic features. The odds of probands with mood-incongruent psychosis having affected first-degree relatives with mood-incongruent psychosis were compared with the odds of probands without this feature having such relatives. A second analysis was restricted to those subjects with psychosis only, and the odds of having a relative affected with mood-incongruent psychosis were compared between probands with mood-incongruent psychosis and probands with mood-congruent psychosis. Odds ratios, controlled for sex, age and duration of illness, were calculated in STATA using logistic regression and generalized estimating equations, which take into account potential correlation between observations when multiple members of the same family are analyzed (25) .
Linkage analyses
We analyzed the genotype data using an affected relative pair approach implemented in the program LODPAL of the S.A.G.E. package (version 4.4). This approach is based on a conditional logistic parameterization of the recurrence risk ratios for offspring and monozygotic twins, conditional on any number of covariates (26) . The model provides estimates for a parameter that measures the “average” linkage in the cohort as well as for parameters that measure the change in linkage as a function of the covariates. Two phenotypic models were evaluated: 1) a “broad” model consisting of at least one sibling with bipolar I disorder; schizoaffective disorder, bipolar type; bipolar II disorder; or recurrent major depression and 2) a “narrow” model excluding recurrent major depression.
The effect of mood-incongruence on linkage was examined by comparing models with and without mood-incongruence entered as a covariate. Results are reported as logarithm of the odds ratio (LOD) scores, derived by dividing the likelihood ratio statistic from the fitted model by 2log e 10. Significance values were derived based on the asymptotic distribution of the likelihood ratio statistic under the null hypothesis of no linkage. The distribution of the likelihood ratio statistic for the baseline model with no covariates is a 50:50 mixture of a point mass at zero and chi square distribution with one degree of freedom. The distribution of the likelihood ratio statistic for the one covariate model is a 50:50 mixture of chi square distribution with one degree of freedom and two degrees of freedom. To test the significance of the covariate effect, the likelihood ratio statistic difference between models with and without the covariate was compared with a chi square distribution with one degree of freedom.
Since asymptotic p values may be liberal, we evaluated the significance of our best findings by randomly simulating genotypes based on our families and pattern of missing data 10,000 times using MERLIN. The number of replicates with a LOD score across the chromosome as high as the original result was divided by 10,000 in order to yield the empirical chromosome-wide p value for overall linkage. To test the empirical significance of the change in LOD score (covariate effect) with mood-incongruence, we randomly permuted the mood-incongruence covariate values among the affected relative pairs 10,000 times. The number of times that a replicate had a higher LOD score across the chromosome compared with the observed data was divided by 10,000 in order to yield the empirical chromosome-wide p value for the covariate effect. Genome-wide significance levels were obtained by multiplying the chromosome-wide p value by a correction factor derived from dividing the genome length in centimorgans (3,500 cM) by the length of the simulated chromosome.
Results
From the 3,643 subjects in the clinical dataset, 2,246 subjects were diagnosed with a major mood disorder: 1,561 (70.1%) had bipolar I disorder; 253 (10.6%) had bipolar II disorder; 348 (15.5%) had recurrent major depression; and 84 (3.7%) had at least one sibling with either bipolar I disorder or schizoaffective disorder, bipolar type. Among all subjects, 799 had psychotic symptoms (35.6%), and of these, 359 (44.9%) were classified as having mood-incongruent psychotic features. The cohort with mood-incongruent psychotic features consisted of 291 subjects with bipolar I disorder (81.1%), three with bipolar II disorder (0.8%), six with recurrent major depression (1.7%), and 59 with schizoaffective disorder, bipolar type (16.4%). No significant differences in sex (63.1% women) or race (92.8% Caucasian) were seen between subjects with and without mood-incongruent psychotic features. Compared with other affected subjects, subjects with mood-incongruent psychotic features were younger (39.6 versus 43.0 years, t=5.08, df=590, p<0.00005) and had a slightly shorter duration of illness (20.5 versus 21.9 years, t=2.01, df=544, p=0.05).
Clinical Correlates in Bipolar I Disorder Subjects
Bipolar I disorder subjects with mood-incongruent psychotic features were significantly different from both the mood-congruent psychotic bipolar I disorder group and the nonpsychotic group on several measures of illness severity ( Table 1 ), since they were more likely to have been hospitalized, to have attempted suicide, and to have had a history of substance abuse or dependence. The following are measures of illness severity between the mood-incongruent psychotic bipolar I group, mood-congruent psychotic bipolar I group, and nonpsychotic group, respectively: hospitalizations, 89.7%, 82.6%, 69.0%, respectively; suicide attempts 45.4%, 37.4%, 34.3%, respectively; and history of substance abuse or dependence, 36.1%, 27.5%, 22.5%, respectively. Subjects in the mood-incongruent psychotic group also had lower Global Assessment Scale scores than those in the mood-congruent psychotic and nonpsychotic groups (63.5, 69.3, 68.5, respectively). Finally, relative to subjects with mood-congruent psychotic features, subjects with mood-incongruent psychotic features had a higher lifetime prevalence of auditory (55.7% versus 32.7%) and visual (50.5% versus 29.2%) hallucinations as well as persecutory delusions (68.7% versus 46.0%, respectively).
Familial Aggregation
The familial aggregation analysis was first carried out on the complete cohort of 708 probands and 1,224 first-degree relatives with a major mood disorder. As shown in Table 2 , probands with mood-incongruent psychotic features were significantly more likely to have relatives with mood-incongruent psychosis than probands without these features (21.5% versus 12.0%; odds ratio=2.07, p<0.0005). When the analysis was restricted to bipolar I disorder first-degree relatives (N=748), the degree of aggregation increased (29.8% versus 14.4%; odds ratio=2.60, p<0.0005).
To distinguish the specific aggregation of mood-incongruent psychotic features from the aggregation of psychotic features in general, aggregation analyses were restricted to subjects with psychosis only. In these analyses, probands with mood-incongruent psychotic features were significantly more likely to have relatives with mood-incongruent psychosis than were probands with mood-congruent psychotic features. The odds ratios were 2.00 (p=0.004) in the major mood disorder relatives cohort and 2.14 (p=0.004) in the bipolar I disorder only cohort ( Table 2 ).
Linkage
Of the 2,899 genotyped subjects, 2,034 had a major mood disorder, providing 1,435 affected relative pairs in the narrow phenotypic model and 1,954 pairs in the broad model. Of those genotyped and diagnosed with a major mood disorder, 322 had mood-incongruent psychotic symptoms. There were 73 affected relative pairs concordant for mood-incongruent psychotic symptoms in each of the two phenotype models.
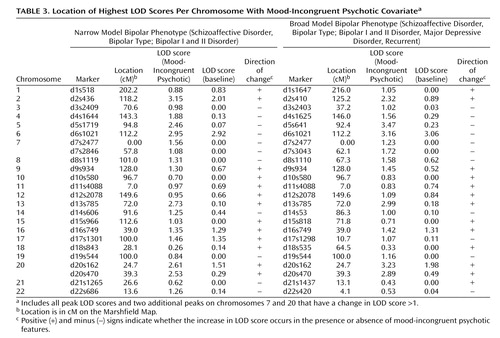
The results of the LODPAL (S.A.G.E.) program analysis with the mood-incongruent psychotic covariate are summarized in Table 3 , which shows the highest LOD scores per chromosome for both phenotypic models and two additional peaks where the LOD score changed by >1 unit. The full covariate genome scan is shown in Figures 1 and 2 . The largest change in LOD score in the presence of the mood-incongruent psychotic covariate was seen on chromosome 13q21-33 in the broad model where the peak LOD at marker d13s785 increased from a baseline value of 0.18 to an LOD of 2.99 (overall LOD nominal p<0.0006, change in LOD nominal p<0.0003). Permutation tests at this locus showed an empirical genome-wide p=0.375 for the overall LOD and p=0.142 for the change in LOD. The 2-LOD support interval under this model spanned 46 cM ( Figure 3 ).
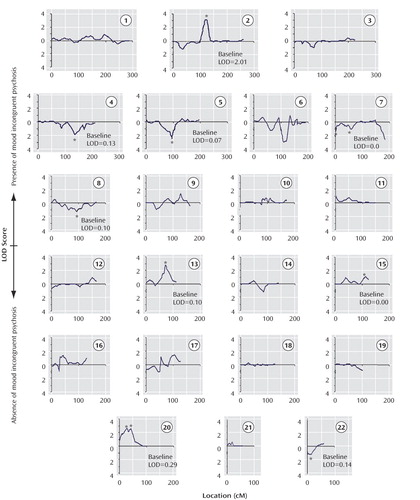
a Changes from baseline >1-LOD are indicated by an asterisk. The y-axis shows positive LOD scores with the upper scale reflecting evidence for linkage in the presence of a history of mood incongruent psychosis and the lower scale reflecting evidence for linkage in the absence of a history of mood incongruent psychosis. The x-axis shows the location along the chromosome in centimorgans (cM) according to the Marshfield map. The baseline LOD score refers to the score obtained with the standard phenotype of schizoaffective disorder, bipolar type; bipolar I disorder; bipolar II disorder; or recurrent major depression.
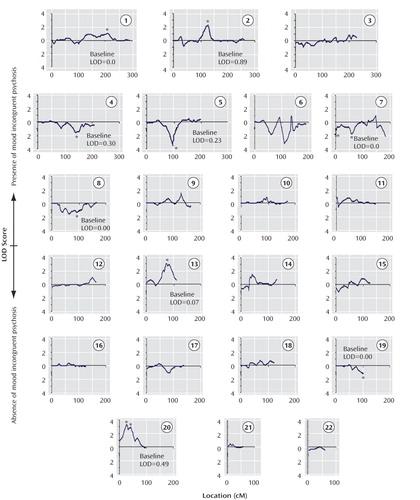
a Changes from baseline >1-LOD are indicated by an asterisk. The y-axis shows positive LOD scores with the upper scale reflecting evidence for linkage in the presence of a history of mood incongruent psychosis and the lower scale reflecting evidence for linkage in the absence of a history of mood incongruent psychosis. The x-axis shows the location along the chromosome in centimorgans (cM) according to the Marshfield map. The baseline LOD score refers to the score obtained with the standard phenotype of schizoaffective disorder, bipolar type; bipolar I disorder; bipolar II disorder; or recurrent major depression.
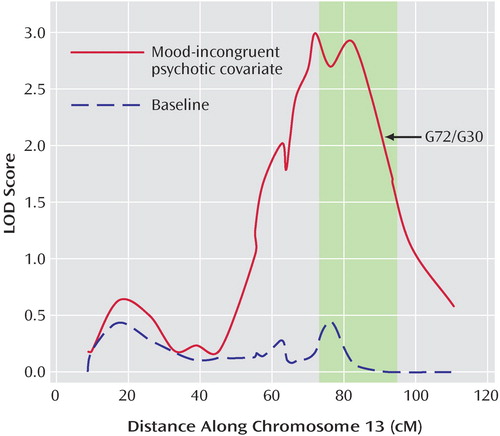
a Shaded region represents previously found overlap regions between schizophrenia and bipolar disorder (27), and arrow shows location of the G72/G30 gene. Location in cM is based on the Marshfield Map.
The highest overall LOD score in the narrow model was found at marker d2s436 on chromosome 2p11-q14, with a LOD of 3.15 in the presence of the mood-incongruent psychotic covariate, compared with a baseline LOD of 2.01 (overall LOD nominal p<0.0004, change in LOD nominal p<0.02). Permutation tests at this locus showed an empirical genome-wide p=0.23 for the overall LOD and p=0.55 for the change in LOD. The 2-LOD support interval spanned 25 cM ( Figure 4 ).
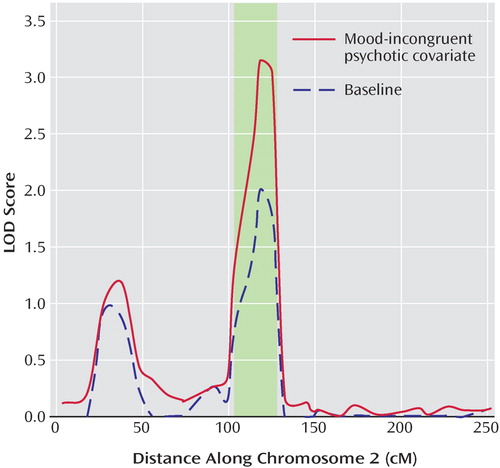
a Shaded region represents findings from a schizophrenia meta-analysis (37). Location in cM is based on the Marshfield Map.
An additional peak in the broad model was seen on chromosome 20p12, with a covariate LOD score of 3.23 at marker d20s162, relative to a baseline LOD of 1.98 (overall LOD nominal p<0.0004, change in LOD nominal p<0.02). Permutation tests of the overall LOD showed a genome-wide empirical p=0.249; however, the change in LOD did not meet genome-wide suggestive linkage criteria, since the empirical genome-wide p=1.11 indicates a result that would be expected more than once per genome scan. A final peak was also seen on 5q14, where the LOD increased from a baseline of 0.23 to 3.47 at marker d5s1719 (overall LOD nominal p=0.0002, change in LOD nominal p=0.0001), but here, the evidence of linkage was strongest among subjects without mood-incongruent psychotic features.
To test the specificity of the 2q and 13q findings, linkage was reexamined in these regions using general psychosis (mood-incongruent psychotic and mood-congruent psychotic) as a covariate. A total of 871 genotyped subjects had a major mood disorder with psychotic features, and there were 261 affected relative pairs concordant for these features. In this larger cohort, the peak overall LOD scores in the identified regions were notably lower than when mood-incongruent psychosis was used as a covariate (2.01 on 2q12 and 1.12 on 13q32).
As a further test of specificity, we evaluated whether persecutory delusions in particular would increase the linkage signal on chromosome 13. There were 519 subjects with a lifetime history of persecutory delusions, of whom, 252 (48.6%) also had mood-incongruent psychotic features. Conversely, 252 (70.2%) of all mood-incongruent psychotic subjects had persecutory delusions. When persecutory delusions were used as a covariate, the highest LOD score obtained was 1.23 at 81 cM (d13s793).
Discussion
In this large cohort of multiplex bipolar disorder families, mood-incongruent psychotic symptoms correlated with a more severe outcome, aggregated in families, and uncovered linkage signals in chromosomal regions previously implicated in schizophrenia. These findings are consistent with the hypothesis that mood-incongruent psychotic symptoms in bipolar disorder may be phenotypic manifestations of genes shared between bipolar disorder and schizophrenia.
This study had several strengths. To our knowledge, the cohort is the largest bipolar disorder family dataset reported to date. Detailed clinical information was gathered using a rigorous and well-validated instrument, and the data were reviewed by data managers and clinicians to minimize information bias. Our results are consistent with most prior studies in showing that subjects with mood-incongruent psychosis had a more severe clinical course than subjects with mood-congruent psychosis. Familial aggregation of mood-incongruent psychotic features was robust, since it was statistically significant under several alternative sample definitions, including those controlling for the clustering of psychotic symptoms in general. Perhaps the strongest validation of our approach is that the two strongest linkage peaks in our analysis were both in regions of a priori significance.
Chromosome 13q31-33 has been the region with arguably the strongest evidence for harboring a bipolar disorder/schizophrenia overlap gene. A meta-analysis identified chromosome 13q31-33 as one of the two strongest linkage regions for both bipolar disorder and schizophrenia (27) , although subsequent meta-analyses differed in their findings (28 , 29) . On 13q32, there have been two genome-wide significant findings reported for schizophrenia (30 , 31) and two suggestive findings for bipolar disorder (32 , 33) . Furthermore, two studies have reported evidence of linkage for psychotic bipolar disorder in this region (19 , 34) , and another showed similar evidence in schizophrenia families with psychotic mood disorder relatives (35) .
The 2-LOD interval for our chromosome 13 linkage peak extends from 55–101 cM on the Marshfield Map (71.0–108.9 Mb on the UCSC genome browser, build 35) and includes the G72(DAOA)/G30 gene complex at 90.6 cM. G72 has been associated with schizophrenia in seven cohorts and with bipolar disorder in five (17) . Schulze et al. examined the G72 gene in two bipolar disorder cohorts and, in the context of examining psychosis in general as well as individual psychotic symptoms, found an association specifically with persecutory delusions (36) . However, they did not examine mood-incongruence. We found that linkage using the persecutory delusions covariate increased the evidence of linkage on chromosome 13q21-33, but not to the extent that the mood-incongruent psychotic covariate did.
Chromosome 2p11-q14 has never been previously implicated in bipolar disorder. However, in the largest schizophrenia linkage meta-analysis, the 2p11-q14 region was the strongest in the genome and the only one meeting criteria for genome-wide significance (37) . Our finding, which peaks at 118 cM on the Marshfield Map, is squarely in the middle of the meta-analytic region of 102–128 cM, and our 2-LOD interval of 105–130 cM (84.0–122.5 Mb) is virtually identical to that region. This region harbors a large number of CNS expressed genes, including the intriguing gene EN1 or Engrailed 1, a homeodomain transcription factor essential for the development and maintenance of midbrain dopaminergic neurons (38) . Given the well-established role of dopamine in psychotic symptoms, both in schizophrenia and bipolar disorder (39) , this gene is worthy of further study.
Our study should be interpreted in light of several limitations. First, although the reliability of mood-incongruence has been established in other studies, we were not able to assess it in our dataset. Second, in light of a conservative statistical assessment employing an empirical genome-wide correction, our results for chromosomes 2p11-q14 and 13q21-33 should be interpreted as suggestive rather than significant evidence for linkage (40) . This is reflected in the genome-wide empirical p values that lie between 0.05 and 1.0, which imply that findings of this magnitude should occur by chance less than once per genome scan. While this level of statistical evidence for the 2p and 13q regions is modest when considered in isolation, it becomes more compelling in light of the converging data referred to previously. Third, for the LODPAL analyses, the number of mood-incongruent psychotic affected relative pairs was small. This may have limited our power to detect loci of modest effect and may explain why we found no linkage evidence for other potential bipolar disorder/schizophrenia overlap regions such as 1q42 (containing DISC1), 6q22, 8p21-23 (containing neuregulin1), 18p11, or 22q11-13.
In conclusion, our results support the validity of mood-incongruent psychosis as a subset of bipolar disorder with increased clinical severity and closer ties to putative psychosis vulnerability genes shared with schizophrenia. The chromosome 13q21-33 finding supports prior evidence of bipolar disorder/schizophrenia overlap in this region, while the 2p11-q14 finding is the first to suggest that this schizophrenia linkage region may also harbor a bipolar disorder susceptibility gene. Use of mood-incongruent psychotic features should inform future linkage and association studies aimed at uncovering genes that confer common susceptibility to these disorders.

1. Jaspers K: General Psychopathology. Johns Hopkins University Press, 1913Google Scholar
2. Black DW, Nasrallah A: Hallucinations and delusions in 1,715 patients with unipolar and bipolar affective disorders. Psychopathology 1989; 22:28–34Google Scholar
3. Azorin JM, Akiskal H, Hantouchec E: The mood-instability hypothesis in the origin of mood-congruent versus mood-incongruent psychotic distinction in mania: validation in a French national study of 1090 patients. J Affect Disord 2006; 96:215–223Google Scholar
4. Andreasen NC, McDonald-Scott P, Grove WM, Keller MB, Shapiro RW, Hirschfeld RM: Assessment of reliability in multicenter collaborative research with a videotape approach. Am J Psychiatry 1982; 139:876–882Google Scholar
5. Tohen M, Tsuang MT, Goodwin DC: Prediction of outcome in mania by mood-congruent or mood-incongruent psychotic features. Am J Psychiatry 1992; 149:1580–1584Google Scholar
6. Craddock N, Jones I, Kirov G, Jones L: The Bipolar Affective Disorder Dimension Scale (BADDS)–a dimensional scale for rating lifetime psychopathology in bipolar spectrum disorders. BMC Psychiatry 2004; 4:19Google Scholar
7. Harrow M, Grossman LS, Herbener ES, Davies EW: Ten-year outcome: patients with schizoaffective disorders, schizophrenia, affective disorders and mood-incongruent psychotic symptoms. Br J Psychiatry 2000; 177:421–426Google Scholar
8. Maj M, Pirozzi R, Bartoli L, Magliano L: Long-term outcome of lithium prophylaxis in bipolar disorder with mood-incongruent psychotic features: a prospective study. J Affect Disord 2002; 71:195–198Google Scholar
9. Toni C, Perugi G, Mata B, Madaro D, Maremmani I, Akiskal HS: Is mood-incongruent manic psychosis a distinct subtype? Eur Arch Psychiatry Clin Neurosci 2001; 251:12–17Google Scholar
10. Coryell W, Tsuang MT, McDaniel J: Psychotic features in major depression: is mood congruence important? J Affect Disord 1982; 4:227–236Google Scholar
11. Coryell W, Tsuang MT: Major depression with mood-congruent or mood-incongruent psychotic features: outcome after 40 years. Am J Psychiatry 1985; 142:479–482Google Scholar
12. Maier W, Lichtermann D, Minges J, Heun R, Hallmayer J, Benkert O: Schizoaffective disorder and affective disorders with mood-incongruent psychotic features: keep separate or combine? Evidence from a family study. Am J Psychiatry 1992; 149:1666–1673Google Scholar
13. Kendler KS, McGuire M, Gruenberg AM, O’Hare A, Spellman M, Walsh D: The Roscommon Family Study, IV: affective illness, anxiety disorders, and alcoholism in relatives. Arch Gen Psychiatry 1993; 50:952–960Google Scholar
14. Potash JB, Willour VL, Chiu YF, Simpson SG, MacKinnon DF, Pearlson GD, DePaulo JR Jr, McInnis MG: The familial aggregation of psychotic symptoms in bipolar disorder pedigrees. Am J Psychiatry 2001; 158:1258–1264Google Scholar
15. O’Mahony E, Corvin A, O’Connell R, Comerford C, Larsen B, Jones R, McCandless F, Kirov G, Cardno AG, Craddock N, Gill M: Sibling pairs with affective disorders: resemblance of demographic and clinical features. Psychol Med 2002; 32:55–61Google Scholar
16. Craddock N, O’Donovan MC, Owen MJ: The genetics of schizophrenia and bipolar disorder: dissecting psychosis. J Med Genet 2005; 42:193–204Google Scholar
17. Detera-Wadleigh SD, McMahon FJ: G72/G30 in schizophrenia and bipolar disorder: review and meta-analysis. Biol Psychiatry 2006; 60:106–114Google Scholar
18. Hodgkinson CA, Goldman D, Jaeger J, Persaud S, Kane JM, Lipsky RH, Malhotra AK: Disrupted in schizophrenia 1 (DISC1): association with schizophrenia, schizoaffective disorder, and bipolar disorder. Am J Hum Genet 2004; 75:862–872Google Scholar
19. Potash JB, Zandi PP, Willour VL, Lan TH, Huo Y, Avramopoulos D, Shugart YY, MacKinnon DF, Simpson SG, McMahon FJ, DePaulo JR Jr, McInnis MG: Suggestive linkage to chromosomal regions 13q31 and 22q12 in families with psychotic bipolar disorder. Am J Psychiatry 2003; 160:680–686Google Scholar
20. Raybould R, Green EK, MacGregor S, Gordon-Smith K, Heron J, Hyde S, Caesar S, Nikolov I, Williams N, Jones L, O’Donovan MC, Owen MJ, Jones I, Kirov G, Craddock N: Bipolar disorder and polymorphisms in the dysbindin gene (DTNBP1). Biol Psychiatry 2005; 57:696–701Google Scholar
21. Green EK, Raybould R, Macgregor S, Gordon-Smith K, Heron J, Hyde S, Grozeva D, Hamshere M, Williams N, Owen MJ, O’Donovan MC, Jones L, Jones I, Kirov G, Craddock N: Operation of the schizophrenia susceptibility gene, neuregulin 1, across traditional diagnostic boundaries to increase risk for bipolar disorder. Arch Gen Psychiatry 2005; 62:642–648Google Scholar
22. Nurnberger JI Jr, DePaulo JR, Gershon ES, Reich T, Blehar MC, edenberg HJ, Foroud T, Miller M, Bowman E, Mayeda A, Rau NL, Smiley C, Conneally PM, McMahon F, Meyers D, Simpson S, McInnis M, Stine OC, Detera-Wadleigh S, Goldin L, Guroff J, Maxwell E, Kazuba D, Gejman PV, Badner J, Sanders A, Rice J, Bierut L, Goate A: Genomic survey of bipolar illness in the NIMH genetics initiative pedigrees: a preliminary report. Am J Med Genet B Neuropsychiatr Genet 1997; 74:227–237Google Scholar
23. Nurnberger JI Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T: Diagnostic Interview for Genetic Studies: rationale, unique features, and training (NIMH Genetics Initiative). Arch Gen Psychiatry 1994; 51:849–859Google Scholar
24. McInnis MG, Dick DM, Willour VL, Avramopoulos D, MacKinnon DF, Simpson SG, Potash JB, Edenberg HJ, Bowman ES, McMahon FJ, Smiley C, Chellis JL, Huo Y, Diggs T, Meyer ET, Miller M, Matteini AT, Rau NL, DePaulo JR, Gershon ES, Badner JA, Rice JP, Goate AM, Detera-Wadleigh SD, Nurnberger JI, Reich T, Zandi PP, Foroud TM: Genome-wide scan and conditional analysis in bipolar disorder: evidence for genomic interaction in the National Institute of Mental Health Genetics Initiative Bipolar Pedigrees. Biol Psychiatry 2003; 54:1265–1273Google Scholar
25. Zeger SL, Liang KY: Longitudinal data analysis for discrete and continuous outcomes. Biometrics 1986; 42:121–130Google Scholar
26. Goddard KA, Witte JS, Suarez BK, Catalona WJ, Olson JM: Model-free linkage analysis with covariates confirms linkage of prostate cancer to chromosomes 1 and 4. Am J Hum Genet 2001; 68:1197–1206Google Scholar
27. Badner JA, Gershon ES: Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry 2002; 7:405–411Google Scholar
28. Segurado R, Detera-Wadleigh SD, Levinson DF, Lewis CM, Gill M, Nurnberger JI Jr, Craddock N, DePaulo JR, Baron M, Gershon ES, Ekholm J, Cichon S, Turecki G, Claes S, Kelsoe JR, Schofield PR, Badenhop RF, Morissette J, Coon H, Blackwood D, McInnes LA, Foroud T, Edenberg HJ, Reich T, Rice JP, Goate A, McInnis MG, McMahon FJ, Badner JA, Goldin LR, Bennett P, Willour VL, Zandi PP, Liu J, Gilliam C, Juo SH, Berrettini WH, Yoshikawa T, Peltonen L, Lonnqvist J, Nothen MM, Schumacher J, Windemuth C, Rietschel M, Propping P, Maier W, Alda M, Grof P, Rouleau GA, Del-Favero J, Van Broeckhoven C, Mendlewicz J, Adolfsson R, Spence MA, Luebbert H, Adams LJ, Donald JA, Mitchell PB, Barden N, Shink E, Byerley W, Muir W, Visscher PM, Macgregor S, Gurling H, Kalsi G, McQuillin A, Escamilla MA, Reus VI, Leon P, Freimer NB, Ewald H, Kruse TA, Mors O, Radhakrishna U, Blouin JL, Antonarakis SE, Akarsu N: Genome scan meta-analysis of schizophrenia and bipolar disorder, part III: bipolar disorder. Am J Hum Genet 2003; 73:49–62Google Scholar
29. McQueen MB, Devlin B, Faraone SV, Nimgaonkar VL, Sklar P, Smoller JW, Abou Jamra R, Albus M, Bacanu SA, Baron M, Barrett TB, Berrettini W, Blacker D, Byerley W, Cichon S, Coryell W, Craddock N, Daly MJ, Depaulo JR, Edenberg HJ, Foroud T, Gill M, Gilliam TC, Hamshere M, Jones I, Jones L, Juo SH, Kelsoe JR, Lambert D, Lange C, Lerer B, Liu J, Maier W, Mackinnon JD, McInnis MG, McMahon FJ, Murphy DL, Nothen MM, Nurnberger JI, Pato CN, Pato MT, Potash JB, Propping P, Pulver AE, Rice JP, Rietschel M, Scheftner W, Schumacher J, Segurado R, Van Steen K, Xie W, Zandi PP, Laird NM: Combined analysis from eleven linkage studies of bipolar disorder provides strong evidence of susceptibility loci on chromosomes 6q and 8q. Am J Hum Genet 2005; 77:582–595Google Scholar
30. Blouin JL, Dombroski BA, Nath SK, Lasseter VK, Wolyniec PS, Nestadt G, Thornquist M, Ullrich G, McGrath J, Kasch L, Lamacz M, Thomas MG, Gehrig C, Radhakrishna U, Snyder SE, Balk KG, Neufeld K, Swartz KL, DeMarchi N, Papadimitriou GN, Dikeos DG, Stefanis CN, Chakravarti A, Childs B, Housman DE, Kazazian HH, Antonarakis S, Pulver AE: Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21. Nat Genet 1998; 20:70–73Google Scholar
31. Brzustowicz LM, Honer WG, Chow EW, Little D, Hogan J, Hodgkinson K, Bassett AS: Linkage of familial schizophrenia to chromosome 13q32. Am J Hum Genet 1999; 65:1096–1103Google Scholar
32. Detera-Wadleigh SD, Badner JA, Berrettini WH, Yoshikawa T, Goldin LR, Turner G, Rollins DY, Moses T, Sanders AR, Karkera JD, Esterling LE, Zeng J, Ferraro TN, Guroff JJ, Kazuba D, Maxwell ME, Nurnberger JI,Jr, Gershon ES: A high-density genome scan detects evidence for a bipolar-disorder susceptibility locus on 13q32 and other potential loci on 1q32 and 18p11.2. Proc Natl Acad Sci USA 1999; 96:5604–5609Google Scholar
33. Kelsoe JR, Spence MA, Loetscher E, Foguet M, Sadovnick AD, Remick RA, Flodman P, Khristich J, Mroczkowski-Parker Z, Brown JL, Masser D, Ungerleider S, Rapaport MH, Wishart WL, Luebbert H: A genome survey indicates a possible susceptibility locus for bipolar disorder on chromosome 22. Proc Natl Acad Sci USA 2001; 98:585–590Google Scholar
34. Park N, Juo SH, Cheng R, Liu J, Loth JE, Lilliston B, Nee J, Grunn A, Kanyas K, Lerer B, Endicott J, Gilliam TC, Baron M: Linkage analysis of psychosis in bipolar pedigrees suggests novel putative loci for bipolar disorder and shared susceptibility with schizophrenia. Mol Psychiatry 2004; 9:1091–1099Google Scholar
35. Pulver AE, Mulle J, Nestadt G, Swartz KL, Blouin JL, Dombroski B, Liang KY, Housman DE, Kazazian HH, Antonarakis SE, Lasseter VK, Wolyniec PS, Thornquist MH, McGrath JA: Genetic heterogeneity in schizophrenia: stratification of genome scan data using co-segregating related phenotypes. Mol Psychiatry 2000; 5:650–653Google Scholar
36. Schulze TG, Ohlraun S, Czerski PM, Schumacher J, Kassem L, Deschner M, Gross M, Tullius M, Heidmann V, Kovalenko S, Jamra RA, Becker T, Leszczynska-Rodziewicz A, Hauser J, Illig T, Klopp N, Wellek S, Cichon S, Henn FA, McMahon FJ, Maier W, Propping P, Nothen MM, Rietschel M: Genotype-phenotype studies in bipolar disorder showing association between the DAOA/G30 locus and persecutory delusions: a first step toward a molecular genetic classification of psychiatric phenotypes. Am J Psychiatry 2005; 162:2101–2108Google Scholar
37. Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, Williams NM, Schwab SG, Pulver AE, Faraone SV, Brzustowicz LM, Kaufmann CA, Garver DL, Gurling HM, Lindholm E, Coon H, Moises HW, Byerley W, Shaw SH, Mesen A, Sherrington R, O’Neill FA, Walsh D, Kendler KS, Ekelund J, Paunio T, Lonnqvist J, Peltonen L, O’Donovan MC, Owen MJ, Wildenauer DB, Maier W, Nestadt G, Blouin JL, Antonarakis SE, Mowry BJ, Silverman JM, Crowe RR, Cloninger CR, Tsuang MT, Malaspina D, Harkavy-Friedman JM, Svrakic DM, Bassett AS, Holcomb J, Kalsi G, McQuillin A, Brynjolfson J, Sigmundsson T, Petursson H, Jazin E, Zoega T, Helgason T: Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: schizophrenia. Am J Hum Genet 2003; 73:34–48Google Scholar
38. Simon HH, Saueressig H, Wurst W, Goulding MD, O’Leary DD: Fate of midbrain dopaminergic neurons controlled by the engrailed genes. J Neurosci 2001; 21:3126–3134Google Scholar
39. Pearlson GD, Wong DF, Tune LE, Ross CA, Chase GA, Links JM, Dannals RF, Wilson AA, Ravert HT, Wagner HNJ: In vivo D2 dopamine receptor density in psychotic and nonpsychotic patients with bipolar disorder. Arch Gen Psychiatry 1995; 52:471–477Google Scholar
40. Lander E, Kruglyak L: Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 1995; 11:241–247Google Scholar