Translational Neuroscience Approaches to Understanding Autism
Abstract
While autism spectrum disorder affects nearly 2% of children in the United States, little is known with certainty concerning the etiologies and brain systems involved. This is due, in part, to the substantial heterogeneity in the presentation of the core symptoms of autism as well as the great number of co-occurring conditions that are common in autistic individuals. Understanding the neurobiology of autism is further hampered by the limited availability of postmortem brain tissue to determine the cellular and molecular alterations that take place in the autistic brain. Animal models therefore provide great translational value in helping to define the neural systems that constitute the social brain and mediate repetitive behaviors or interests. If they are based on genetic or environmental factors that contribute to autism, organisms from flies to nonhuman primates may serve as models of the neural structure or function of the autistic brain. Ultimately, successful models can also be employed to test the safety and effectiveness of potential therapeutics. This is an overview of the major animal species that are currently used as models of autism, including an appraisal of the advantages and limitations of each.
Autism spectrum disorder (ASD or autism) is a neurodevelopmental condition characterized by alterations in two core domains of behavior (1). The first comprises deficits in social and communication abilities. This can manifest in lack of eye contact during communication with others or limited reciprocal interactions or joint attention. The second domain is the presence of restricted or repetitive patterns of behavior and interests. Symptoms range from repetitive movements to very circumscribed interests. Beyond these core symptoms, autistic individuals commonly have one or more co-occurring conditions. These commonly include intellectual disability, anxiety, epilepsy, sleep disorders, gastrointestinal distress, and others. The heterogeneity in core autism traits and in co-occurring conditions results in an extremely broad spectrum of outcomes that range from the need for lifelong care to having highly successful professional careers. Here, we discuss autism spectrum disorder as a DSM-based diagnosis and use identity-first language, reflecting the preferences of autistic self-advocates (2, 3).
While there is a consensus that the etiology of autism is rooted in prenatal factors, the enormous heterogeneity of the condition makes pinpointing causal mechanisms extremely challenging. There is substantial and increasing evidence that many forms of autism have a genetic underpinning. However, the genetic architecture of autism is also extremely complex. The first genetic associations in autism were with genetic syndromes that often, but not always, include autism traits, such as fragile X syndrome, where ASD is found in ∼30% of affected individuals. More recent findings have been driven primarily by rare mutations (de novo variants) detected within large cohorts of autistic individuals. Currently, at least 185 single genes (4) are known to be associated with autism. Taken together, however, the genetic variants provide explanatory evidence for only about 20% of autism cases. Moreover, none of the identified genetic variants is deterministic, with most associated variants yielding a 10%–50% likelihood of autism diagnosis, whereas the rate in the general population is ∼2% (5). Beyond genetic factors, there is mounting evidence that the immune system may also play a role in the etiology of some forms of autism. For example, there is speculation that maternal antibodies that cross-react with fetal brain tissue may alter brain development (6). Similarly, activation of the maternal immune system through viral or bacterial infection and the resulting cytokine barrage has also been proposed as pathogenic for autism (7).
MRI studies of autistic individuals have demonstrated a variety of alterations in brain structure and function (8–10). Some of the alterations are associated with the core symptoms, while others are more highly related to one of the co-occurring conditions (11). Given the complexity of autism, however, a reasonable expectation is that multiple brain structures and systems will be involved. The limited number of postmortem neuropathological studies of the autistic brain that have been carried out thus far have failed to find a consistent feature that is diagnostic of autism (12). Thus, efforts to appreciate the underlying biology responsible for autistic symptoms has fallen to studies of model systems, with an emphasis on animal models, to connect molecular events to neurodevelopment and behavior.
In this overview, we summarize translational studies carried out in nonmammalian species (mainly drosophila and zebrafish), in rodents (mainly mouse models), and in nonhuman primates (Figure 1). Basic neuroscience studies that attempt to define the neurocircuitry of social or repetitive behavior have contributed greatly to our understanding of the brain systems likely to be involved in autism. Here we focus on work that attempts to create animal models of autism risk factors, most often currently through genetic manipulations, with the explicit intention of understanding the symptoms and biology associated with human autism. Importantly, models, by definition, fail to recapitulate the entirety of the human condition and must be considered carefully for their purpose and utility. There are myriad goals for animal models, ranging from understanding the causal biology of autism to creating preclinical testbeds for evaluating the safety and effectiveness of promising therapeutics. There is by no means a consensus on which animal model system is the most appropriate to achieve these goals. And there are clearly advantages and disadvantages for each. There is also the hope that the use of human induced pluripotent stem cells and resulting organoids may offer new insights, particularly into early stages of human neurodevelopment, while also reducing the number of animals used in preclinical research on autism. We touch on several of these issues in this overview.
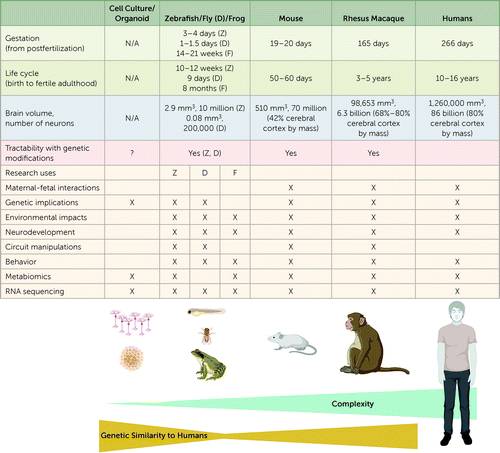
FIGURE 1. Translational models in autism researcha
aMice, monkeys, and humans have the greatest neural complexity, followed by the nonmammalian vertebrates, zebrafish, fruit flies, and frogs. While monkeys and mice have substantial genetic similarity to humans, human organoids have the greatest genetic similarity. Z=Danio rerio, zebrafish; D=Drosophila melanogaster, fruit fly; F=Xenopus tropicalis, frog. (Created with BioRender.com.)
Nonmammalian Models
Invertebrate Models
Due to the genetically heterogeneous nature of autism, studies in nonmammalian models have enabled functional tests of multiple genes at relatively higher throughput compared with traditional model systems. Studies using invertebrates, such as nematodes (Caenorhabditis elegans, reviewed in reference 13) and fruit flies (Drosophila melanogaster, reviewed in references 14, 15), leverage the deep knowledge of neurobiology in these systems, including neurogenesis and synaptogenesis, to understand molecular and developmental impacts of loss-of-function mutations of gene orthologs conserved with humans. Stable mutants are easily generated for both systems, with life cycles on the order of 3 days for nematodes (16) and 9–10 days for fruit flies (17).
The small and transparent nature of C. elegans make this species particularly well suited for imaging-based phenotypic screens, and the developmental trajectory of the 302 neurons comprising their nervous system has been extensively studied over the past 50 years. One product of this effort is a complete connectome map (18). As a result, hundreds of autism candidate genes have been studied in C. elegans, perhaps best represented by a recent study that generated 135 mutant knockout strains and tested their impacts across 26 morphological and behavioral phenotypes, such as locomotion and habituation learning (19).
Similarly, studies in Drosophila have largely taken advantage of conservation of synaptogenesis and synaptic plasticity with humans to study autism candidate genes implicated in these functions (e.g., Shank) (20, 21). In addition to high-confidence autism genes, a more recent study used fruit flies to screen 74 genes with discovered loss-of-function de novo variants with predicted high effect sizes from the Simons Simplex Collection (comprising quad families with unaffected parents, siblings, and a single autistic proband) that lack genetic associations with ASD due to lack of mutational recurrence in patients (22). By generating autistic patient–derived alleles in both fly reference and “humanized” genes and characterizing impacts on varied phenotypes—including lethality, neurodevelopment, and courtship behavior—this screen implicated several previously known as well as novel genes as putatively contributing to ASD.
Vertebrate Models: Highlighting Zebrafish
More complex vertebrate systems, such as zebrafish (Danio rerio, reviewed in reference 23) and frogs (Xenopus laevis and X. tropicalis, reviewed in reference 24), can also efficiently produce genetic mutants across hundreds to thousands of progeny while more closely modeling the mammalian brain and neural networks. Here, we highlight work in zebrafish, as they provide a powerful tool to understand vertebrate development and disease (25). External fertilization, rapid development, embryonic transparency, small size, and powerful available gene-editing tools are some of the advantages that make zebrafish an attractive model for performing genetic screens (26). Zebrafish have a ∼1.4-gigabasepair genome distributed across 25 chromosomes with 26,202 protein-coding genes. At least one obvious ortholog has been identified for ∼70% of human genes (27). This information has enabled creative techniques to use CRISPR-based assays to study single or multiple gene targets, recapitulating previous findings from other model organisms and providing new, valuable information (28).
Although mammalian models, such as mice and primates, have greater genetic and morphological similarity to humans, zebrafish provide a higher-throughput, affordable option to characterize neurodevelopmental traits and disorders (29, 30). Despite notable differences from humans, such as the absence of a neocortex, recent reviews highlight the degree of conservation in the development of the nervous system. This includes a conserved structural organization of major brain divisions, similar gene expression patterns in early brain development, and the use of the same major neurotransmitters (26, 31). The central nervous system forms rapidly in zebrafish, allowing for fast phenotypic assessments of multiple embryos simultaneously. Neurogenesis in zebrafish begins within day 1 of development (16 hours postfertilization), with demarcation of the midbrain-hindbrain boundary, cerebellum, and thalamus. The blood-brain barrier forms by 20 hours postfertilization, and core neural signaling is established between 18 and 32 hours postfertilization (32). Moreover, the resulting neural circuitry has been shown to be conserved between zebrafish and mammals, so gene impacts can be tested at a behavioral level. Recording-based evaluations in zebrafish larvae can be used to detect impairments in startle responses to visual or acoustic stimuli as well as frequency of chemical-induced or spontaneous seizure-like movements (33, 34). In adults, it is possible to test for differences in complex social behaviors, including preferences to conspecifics, shoaling, schooling, aggression, and repetitive behaviors, among many other autism-relevant behaviors (35). Moreover, it is possible to rapidly investigate accompanying autism co-occurring phenotypes in zebrafish mutants, such as changes in sleep patterns (36) and gut-brain-microbiome axis alterations (37). Together, these combined characteristics establish the robustness of zebrafish as a model for the functional interrogation of autism candidate genes.
Clinical neurological phenotypes have been examined in zebrafish studies of autism candidate genes, including cranial malformations observed in autistic patients with chromosome 16p11.2 duplication (38), GABAergic neuron deficiencies due to mutations in the autism risk gene CNTNAP2 (39), activity changes and seizures caused by alterations in SCN1A (40), developmental delay and reduced social behaviors in SHANK3 mutants (41), increased head size and impairment of gastrointestinal motility due to the disruption of autism-associated gene CHD8 (42), and reduced social behaviors in mutants of EGR1 (43), among others in a growing list of examples. More recently, zebrafish have been used in large CRISPR-based screens that couple gene knockouts and high-throughput imaging to detail the impact of genes putatively involved in schizophrenia (44) and synaptogenesis (45). These screens are valuable for identifying candidate genes with functional impacts from the many putative autism risk genes found in human genomic assessments. These candidate genes can then be further studied in complementary animal models (e.g., rodents and primates) to dissect their roles in specific developmental processes.
Rodent Models
Since the initial description of the Fmr1 knockout mouse model of fragile X syndrome (46), rodent models have been the mainstay of research on the cellular, developmental, and circuitry consequences of autism risk factors. Most of this work has been conducted in mice, using an extensive molecular tool kit that allows gene knockout, overexpression, and rescue, including cell-specific and developmental gene targeting, as well as manipulation of individual cells and circuits using optogenetic and chemogenetic tools. With the availability of CRISPR-Cas9 and other flexible gene manipulation approaches, researchers have also incorporated more rat models to harness their greater cognitive abilities. The availability of this technology also allows for studies in less commonly used rodents, such as prairie voles, that may offer additional opportunities for understanding more complex social behavior, such as pair bonding (47).
Unlike in nonmammalian species, where social behavior is so different that direct parallels are rarely drawn to the human, some work in rodents has been focused on the underpinnings of observed behavior or cognition, rather than modeling genetic or environmental risk factors. Before the identification of robust autism risk genes, inbred mice with unusual patterns of social disinterest or repetitive behavior were studied to understand both their patterns of behavior and the potential underlying brain mechanisms (48, 49). This work did not yield clear findings that mapped onto the human condition, which led to substantial caution about models that are based solely on face validity, or similar patterns of behavior between mice and humans. This should not be surprising in the context of both the evolutionary and behavioral differences between humans and rodents, in contrast with nonhuman primate models.
More recently, optogenetic and chemogenetic tools have allowed detailed dissection of circuits underlying behaviors in mice, from social reinforcement to habitual behavior. These “circuit cracking” studies fit into the National Institute of Mental Health Research Domain Criteria approach, which may yield an understanding of constructs such as social reward across species, even without directly connecting to the biology of autism risk. As one example, the Costa lab has focused on cortico-striatal circuits that underlie habit versus goal-directed learning in operant paradigms (50, 51). As a potentially translatable example, the Malenka lab has used a combination of optogenetics and pharmacology to map dorsal raphe projections to serotonin 5-HT1B receptors in the nucleus accumbens that are necessary for social-conditioned place preference in mice (52, 53). These findings yield hypotheses that can then be tested to evaluate relevance to the human condition, such as an ongoing randomized controlled trial of a 5-HT1B agonist in autism (ClinicalTrials.gov identifier: NCT05081245).
With the availability of robust environmental and genetic risk factors in autism, mouse models have been increasingly based not on face validity but on construct validity, or recapitulating the same etiology across species. Some environmental risk factors have been extensively modeled in rodents. For example, prenatal exposure to valproic acid has been modeled based on occurrence of ASD in as many as 15% of offspring, as well as risk of intellectual disability, neural tube defects, and other congenital malformations (54). Various doses and timing of developmental exposure to valproic acid in mice and rats produce a range of behavioral and cognitive deficits (55), with some data pointing to chromatin regulation as one etiological mechanism (56). Rodent models of maternal immune activation have also been extensively studied, given an increased risk of autism following maternal infection or fever during pregnancy, with odds ratios for subsequent autism diagnosis ranging from 1.1 to 1.3 in meta-analyses (57, 58), and larger effect sizes for risk of schizophrenia (59; review in reference 60). Patterson and colleagues demonstrated that maternal infection in mice leads to a cytokine cascade that has deleterious effects on the development of the brain and on behavior (7, 61). Various approaches to maternal immune activation in mice have mimicked either viral or bacterial infection to identify key cytokine pathways, including interleukin-6 and -17, that contribute to subsequent cognitive and behavioral abnormalities (62, 63). In contrast to environmental exposures, genetic risk variants are inherently simpler to model because they can directly parallel disruption of a single gene or chromosomal region, allowing researchers to be more confident that their findings will be relevant to humans.
Construct and face validity do not always overlap, at least not in ways that clinicians might hope. As the oldest example, children with fragile X syndrome have moderate intellectual disability, substantial hyperactivity, and a ∼30% risk of autism, whereas the corresponding mouse model shows only mild hyperactivity, with inconsistent changes in the cognitive and social tasks that most easily map onto human behavior (46, 64). This raises considerable challenges when investigators seek to translate potential treatments from mice to humans, when the behaviors to be rescued don’t match. As noted above, no autism risk gene leads to autistic traits in every case, indicating that there are also other genetic or environmental factors that moderate likelihood of autism. We must also expect variability in social or repetitive behavior across models of autism. Indeed, a heroically complex study from Tabbaa and colleagues found considerable variability in altered social behavior and brain size when crossing a Chd8 heterozygous knockout mouse with 33 different inbred strains of mice (65). Interestingly, they found that the most consistent change was in dominance behavior, a measure of the rodent social hierarchy that does not map well onto assessments of autistic social behavior. This calls into question the application of mouse behavioral tasks that resemble human behaviors and instead suggests a more ethologically valid approach to assessing behaviors that are relevant to mouse social function.
A third approach to model validation, predictive validity, posits that we can be confident that we are studying the same biology when an intervention has a parallel effect across species. Most rodent models applied to depression have therefore been based on behavioral response to serotonin reuptake inhibitors or other drugs with antidepressant effects. Unfortunately, there are currently no treatments for the core symptoms of autism that can be applied across species. Although some investigators have evaluated the impact of risperidone on repetitive grooming behavior in mice (e.g., 66, 67), the indication approved by the U.S. Food and Drug Administration for use of risperidone in autistic people is for difficulties with agitation or mood reactivity. Behavioral interventions that are helpful in humans cannot be directly paralleled in mice, although some investigators have demonstrated that exposure to different groups of peers can change social behavior in mice (68).
With rodent models primarily based on construct validity, researchers can focus more clearly on the goals of using an individual model. In most cases, publications on animal models have been focused on understanding the biology underlying the corresponding risk factor in humans. In the initial models that were developed, this was straightforward, such as fragile X syndrome, where there is complete loss of FMR1 gene expression (64). Even where the gene has substantial homology across species, however, there may be substantial differences in gene expression or protein function across development or across different cellular subtypes. For example, the fragile X protein FMRP regulates the nitric oxide synthase gene in developing neocortex pyramidal cells in primates but not in rodents (69), suggesting that the mouse model of Fmr1 loss may not have construct validity for this aspect of human neocortical development, despite modeling other aspects of human brain development and function. At least at the level of gene expression, we are making strides toward understanding similarities and differences across species to better evaluate the suitability of a model to a given brain region and period of development (70, 71).
The ongoing tension between construct and face validity is most easily seen in choices investigators make when modeling rare gene mutations that have dominant effects. Most autism risk genes follow this pattern, where a single copy of the gene—such as SHANK3, the gene responsible for Phelan-McDermid syndrome—is mutated or deleted, leaving one intact copy. Modeling loss of a single copy of Shank3, however, does not lead to a dramatic phenotype in mice, so the vast majority of studies have instead focused on homozygous mice lacking Shank3 entirely, a condition that has never been found in a human. These animals do show repetitive grooming behavior and inconsistent decreases in sociability (72), but it is not clear how these behavioral changes should be conceptualized in relation to autism. In this context, the heterozygous null mouse seems to be a model of Phelan-McDermid syndrome and of autism risk, whereas the homozygous mutant may still be a useful experimental system for probing the underlying biology. This calls into question, though, the use of the full knockout to develop potential treatments that would be applied to humans who are missing only one copy of the gene (73, 74).
Overall, rodents have many advantages over nonmammalian models at the level of brain development, conserved circuits, and behavior. Many examples have emerged of interventions that improve brain or behavior in rodent models of autism-associated syndromes (75–78). However, none of these has yet delivered new treatments to the clinic, although very few of these hypotheses have been adequately tested. The challenges in translating these findings should not be surprising given the evolutionary distance from humans, as reflected in neuronal and glial complexity, brain size, and behavior. The faster reproductive and developmental time periods, coupled with the available tools for genetic and circuit manipulations as well as the ready availability of the brain for both in vivo and ex vivo studies, still make rodents an ideal system to bridge from cellular and invertebrate models. The gap between rodents and humans can also increasingly be filled by the application of similar tools in nonhuman primate models.
Nonhuman Primates
Arguments for the value of developing nonhuman primate models of human health issues have been cogently made on several occasions (e.g., 79). We briefly summarize some of these arguments, with an initial focus on the most widely used nonhuman primate research species—the macaque monkey (usually Macaca mulatta or M. fascicularis). From an evolutionary perspective, mice diverged from human ancestors around 80 million years ago, whereas macaque monkeys diverged closer to 25 million years ago. Therefore, there has been far greater time for differences in biological and behavioral systems to emerge in mice, which favors the nonhuman primate in translational autism research. This greater similarity to humans makes the nonhuman primate ideal for modeling health-related issues ranging from reproduction to respiratory illness to infectious disease to neurodevelopmental disorders.
Several key factors argue in favor of nonhuman primate models of autism (80, 81). First, and foremost, is the substantially greater homology of the structure and connectivity of brain regions thought to be critical in the etiology of autism. While the brain of the macaque monkey is about 1/10th the size of that of a human, virtually all of the regions identified in the human brain are also seen in the macaque monkey; this is not the case for the mouse. The frontal cortex is an excellent example. The prefrontal cortex is considered to play a key role in mediating social function and appears to be altered in autism (82, 83). There is substantial evidence that the mouse lacks the dorsolateral portion of this cortex, which is prominent both in humans and in macaque monkeys (84, 85). There are also substantial cytoarchitectonic and connectional differences between the mouse frontal cortex and the closest homologs in the primate brain.
Additionally, nonhuman primates, and particularly macaque monkeys, have a behavioral repertoire that more closely replicates that of humans than does the rodents’. Many of these behaviors, such as social interaction and stereotyped behaviors, are the diagnostic features of autism. Mice simply lack the sophisticated social and cognitive abilities of humans. Rhesus monkeys, in contrast, display complex social behaviors. Much like humans, macaques in the wild and in some research facilities spend their lives in complex societies where survival depends on their ability to quickly and accurately interpret and respond to a variety of social signals. Several generations of macaque female relatives live together and form long-lasting social networks or matrilines. These matrilines are organized into dominance hierarchies, where prediction of social rank is closely linked to the dominance status of kin. This complex system of social organization requires an equally sophisticated social communication system that includes interpretation of facial expressions, vocalizations, and body postures. Like humans, but unlike rodents, macaque monkeys use various facial expressions to convey social intent (86, 87). This is particularly germane to models of autism, since autistic humans often have difficulties in evaluating or understanding facial expressions (88, 89). Macaque monkeys also spontaneously develop stereotypies of various types, ranging from whole body (twirling, flipping) to self-directed (eye poking, hair pulling, and self-grasping) (90, 91). The macaque monkey therefore provides a valuable model for studies of the neurobiology of stereotyped behavior and for treatment studies.
Despite the neuroanatomical and behavioral advantages of nonhuman primate models of autism, there are also some clear limitations. First, the enhanced cognitive and social capabilities of nonhuman primates raise ethical issues about their use as animal models. While these are not to be taken lightly, there are cogent arguments for why it is ethical to employ them to reduce human suffering (79). In fact, the Nuremberg Code, an outcome of the Nuremberg trials after World War II, stipulates that research on humans must be preceded by animal studies (92). However, in order to minimize the number of animals used, nonhuman primate studies often rely on very small samples. Even with small sample sizes, nonhuman primate research is inherently costly. Obtaining an adequate number of animal subjects and conforming to regulatory requirements for animal husbandry, veterinary care, and housing is prohibitively costly for many investigators and some academic institutions. Thus, much nonhuman primate research is carried out in specialized primate centers such as the National Primate Centers in the United States and Japan.
Beyond these limitations is the time factor involved in nonhuman primate research. The normal gestation of a rhesus macaque monkey is approximately 165 days. The normal lifespan of a rhesus monkey is approximately 30 years. For developmental research, rhesus monkeys usually have a single offspring; the prevalence of twins is 0.1% (93). Thus, producing genetically modified nonhuman primates and propagating developed lines is far more time-consuming than similar projects with mice. Finally, macaque monkeys are aggressive, and they can harbor the herpes B virus in saliva and bodily fluids, which is potentially deadly to humans.
Macaque Versus Marmoset Models
Before discussing current nonhuman primate translational models of autism, it makes sense to briefly highlight the benefits and disadvantages of the two most widely used model species. In addition to the rhesus monkey (Macaca mulatta—an Old World monkey) described above, the common marmoset (Callithrix jacchus—a New World monkey) has gained increasing acceptance as a research model, particularly for genetic modifications. There are many advantages to using the marmoset. It is small (300 grams for a mature male, compared with 7–10 kilograms for a mature male rhesus monkey). It also has a shorter gestation (144 days), commonly has twin births, and matures to adulthood in 1.5 years (compared with 3–4 years for the rhesus monkey). Preuss (94) has discussed why the choice of marmosets over rhesus monkeys is not straightforward and concludes that both species should be used, when advantageous, for modeling human health issues. A critical issue is that far less neurobiological information is available for the marmoset compared with macaque monkeys.
Nonhuman Primate Models of Autism
We would suggest that for an animal model to provide a valuable contribution to autism research, it must stem from hypotheses that are directly related to the human disorder. It is important to keep in mind that many biological or environmental factors can perturb the brain systems involved in social behavior but may not be germane to the etiology of autism. For example, profound social isolation produces an alteration of social behavior both in nonhuman primates (95) and in young children (96) but is not a cause of autism (97). Therefore, as in the rodent, “construct validity” is critically important in considering the value of nonhuman primate models of autism.
Naturally occurring social variation.
In a typical study of autistic children, an initial stage is to recruit a cohort of participants who have a diagnosis of autism along with a group of age-matched non-autistic individuals. While this approach is typically not feasible in most primate research laboratories, it is possible at the National Primate Centers. This approach has been advocated by Capitanio (98), and more recently by Parker (99), to attempt to identify macaque monkeys with naturally occurring autistic symptoms. Using a revised version of the Monkey Social Responsiveness Scale (100, 101), modeled after a similar instrument for use with humans, it was shown that male rhesus monkeys demonstrate pronounced individual differences in autistic-like traits. This strategy allows the identification of biological measures that differentiate the animals with low and high autistic traits. The researchers demonstrated, for example, that cerebrospinal fluid levels of arginine vasopressin differentiated those animals with higher autistic traits. A similar strategy has been used by Gunter et al. (102) to select macaque monkeys with high autism-like traits and then to genotype them to determine whether there are any variants related to known autism risk genes. This would appear to be a very promising strategy for determining homologies of genetic variants in macaque monkeys and known autistic risk variants. However, for these behaviorally driven strategies to be fully informative, the number of monkeys sampled will need to be increased substantially, perhaps through developing consortia similar to those that have been developed for studying the genetics of human autism.
Valproic acid model.
The valproic acid model of autism has been applied in the marmoset monkey, as in the rodent. A series of papers from the laboratory of Ichinohe (103, 104) have reported behavioral alterations consistent with the core symptoms of autism, as well as alterations in synaptogenesis and gene expression.
Maternal immune activation.
As noted above, there is substantial epidemiological data indicating that maternal infection during pregnancy can increase the likelihood of offspring having a diagnosis of schizophrenia (59) or autism (57, 58). The maternal immune activation model of neurodevelopmental disorder has been evaluated both in marmosets (105) and in rhesus monkeys (106). In both cases, treated offspring demonstrated altered communication and repetitive behaviors as well as indications of abnormal species-typical social behavior (107, 108). In recent work by Vlasova et al. (108), longitudinal MRI revealed significant gray matter volume reductions in the prefrontal and frontal cortices of maternal immune activation–treated offspring at 6 months that persisted through 45 months, along with smaller frontal white matter volumes in treated animals at 36 and 45 months. There are also indications of altered dendritic morphology in maternal immune activation–treated macaques (109). The maternal immune activation model provides an excellent example of the potential for translating findings from rodent to monkey (which were initially based on human epidemiology), with the prospect of identifying protective strategies that may contribute to the prevention or reduction of severity of some neurodevelopmental disorders.
Maternal antibody–associated autism.
Since the early 2000s, researchers have considered the proposal that some forms of autism are caused by exposure of the fetal brain to circulating maternal antibodies that disrupt normal brain development (110). Van de Water and colleagues have identified several antibodies that are largely unique in up to 20% of women who have given birth to autistic children (111). Because placental physiology is substantially different in primates compared with rodents, it was important to determine whether these antibodies, when injected into pregnant rhesus monkeys, would produce offspring with altered behavior. In an initial study by Martin et al. (112), rhesus monkeys gestationally exposed to IgG class antibodies derived from mothers of autistic children consistently demonstrated increased whole-body stereotypies across multiple testing paradigms. This was not the case for offspring of mothers injected with IgG from mothers of non-autistic children. In a follow-up study with more selective antibody treatments, Bauman et al. (113) also detected both behavioral and brain alterations. Behavioral differences were first detected when the macaque mothers responded to their exposed offspring with heightened protectiveness during early development. As they matured, offspring exposed to antibodies from mothers of autistic children consistently deviated from species-typical social norms by more frequently approaching familiar peers. Even more striking, these offspring displayed inappropriate approach to unfamiliar peers, clearly deviating from normal macaque social behavior. Longitudinal MRI analyses revealed that male offspring had enlarged brain volume compared with control monkeys, due largely to white matter volume increases. While these observations have excellent construct validity and raise opportunities for intervention (by blocking the antibodies during pregnancy) the current studies have several limitations. Foremost among these is that the IgG exposure takes place during a short portion of the pregnancy, whereas in the human condition, antibodies potentially have access to the fetal brain during much of pregnancy. This model would be strengthened if the rhesus females could be induced to generate similar antibodies for the full gestational period.
Genetically modified monkeys.
The development of highly efficient gene-editing systems such as CRISPR-Cas9 has opened the potential for modeling human disorders such as autism by targeting genes that are highly associated with the condition. While numerous rodent models have been produced that target autism susceptibility genes, the variability in, and often lack of, phenotypes resembling autistic symptoms has motivated the field to consider using other model systems, including nonhuman primates (80, 114, 115). This strategy is particularly enticing given the recent improvements in sequencing of the rhesus monkey genome (116). The promise of genetic engineering of nonhuman primates for translational purposes has been reviewed previously (e.g., 114), as well as for producing models of autism (81). Initial neurodevelopmental gene-editing studies utilized TALEN (transcription activator-like effector nuclease) in macaque monkeys to target MECP2, the gene involved in Rett syndrome (117).
Published studies on modeling autism in macaque monkeys have focused on editing of SHANK3 using CRISPR-Cas9 technology. Mutations of SHANK3 have been identified in many individuals diagnosed with autism and are considered a high-confidence causal factor. SHANK3 is located in a multigenic region of chromosome 22 that is deleted in Phelan-McDermid syndrome, which is frequently accompanied by a diagnosis of autism. Initial attempts at producing a macaque model of SHANK3-related autism were reported by Zhao et al. (118). The investigators obtained one live birth after implanting 116 embryos in 37 surrogate mothers (the low production of viable offspring is a limitation of gene-edited nonhuman primates, which remains a challenge). A second research group is also producing SHANK3-mutant macaque monkeys (119). This group produced four living males and one living female with SHANK3 mutations, and all of them had decreased brain protein products of the gene. The mutant monkeys shared many characteristics with patients with Phelan-McDermid syndrome, including sleep disturbances, increases in stereotyped behaviors, and hypotonia. They also demonstrated altered species-typical social behavior, including changes in eye gaze behaviors. MRI studies revealed decreases in gray matter. Analysis of functional connectivity suggested long-range hypoconnectivity but short-fiber-connection hyperconnectivity. In summary, many of the characteristics of these mutant macaque monkeys more clearly map onto phenotypical expectations than has been found with heterozygous mouse models of Phelan-McDermid syndrome. While genetically modified monkey models of autism remain attractive for evaluating the neurobiology of autism, the field is at a very preliminary stage, and whether the potential will be realized remains to be determined.
Discussion: Animal Models in Context
Animal models offer the potential for developing and testing hypotheses before attempted translation to human populations. Initial studies in most model systems seek to identify potential mechanisms by paralleling known factors that contribute to autism likelihood in human populations. Various animals, from worms to primates, offer quite different advantages for these studies, with gradients of cost, throughput, and complexity (Figure 1), along with available tools, driving selection of the ideal model for the specific questions targeted. In the context of substantial limitations in studies of the living brain in humans, animal models offer opportunities to study molecular and cellular cascades across development that are likely to remain impactful in autism research for the foreseeable future. The limitations of animal systems for modeling a human condition must also be carefully considered, including substantial differences in brain size and complexity, as well as the impossibility of paralleling human social behavior in the absence of language.
One alternative to animals as a model of the human brain is the use of induced pluripotent stem cell (iPSC) preparations. These potentially offer obvious advantages over animal models at the level of genomic identity, gene regulation, and patterns of protein expression across development. Importantly, iPSC-derived neurons and glia may overcome the limitation inherent to models of genetic or environmental factors that are not deterministic for autism in the human population. Even the most well-characterized genetic syndromes associated with autism lead to a diagnosis in only a minority of affected individuals (120). In contrast, iPSCs can be directly derived from autistic people, including those without known genetic or environmental risk factors (121). Animal models cannot achieve construct validity for so-called idiopathic autism, which lacks a known major risk factor.
Initial work with iPSC-derived neurons and glia used cell-culture or two-dimensional approaches, which offer opportunities to evaluate gene expression or synaptic connections. Increasingly, three-dimensional cerebral organoids are an option for studying the patterns of brain development and connections that develop between neurons over time (122). By combining separately developing organoids into composite “assembloids,” it is possible to model the infiltration of interneurons into the developing cortex (123) or developing connections between cortex and striatum (124). Like animal models, these iPSC-derived models have inherent limitations, most prominently the lack of a behavioral readout to evaluate whether observed molecular and cellular changes, or potential interventions, are relevant to autistic traits in humans. Cerebral organoids also primarily model early regional brain development. Although recent data indicate molecular transitions in iPSC-derived cerebral organoids that extend beyond the prenatal period and into early postnatal development (125), they fall well short of the periods of human brain development when a diagnosis of autism might be made. With convergent genetic and environmental data pointing to fetal development as the key period for autism risk (126–128), organoids and other iPSC preparations may be well suited for understanding the molecular cascades of autism risk but are unlikely to parallel the time points when intervention may be possible in humans. There may be opportunities to bridge iPSCs into whole-animal preparations, however. Earlier work had already shown that human iPSC-derived neurons could be integrated into rodent brains (129), and recent data indicate that organoids can also be incorporated, allowing assessment of their impact on functional brain circuits and behavior (130), although the much larger size of human neurons suggests that human-to-rodent transplants may meet challenges in assessing complex circuits and behavior.
Regardless of advances in animal and cellular model systems, the emergence of gene-based treatments may short-circuit the need to understand all the downstream neurobiological consequences that are typically investigated in animal models. In spinal muscular atrophy, for example, intrathecal infusion of antisense oligonucleotides has revolutionized care, with improvements in motor development and in lifespan for treated children. CRISPR-Cas9–based treatments potentially offer an even more direct benefit, whether targeting genetic variants directly or manipulating gene expression via epigenetics. The risks associated with gene-based therapies suggest that they should be reserved, at least initially, for situations in which outcomes can be predicted with confidence to include profound neurological impairment or premature death, and initial work is already under way in Angelman syndrome and Rett syndrome. We are not far from a future where genome sequencing will be used as a prenatal or newborn screen. Although this will provide opportunities for prediagnostic gene-based interventions, we will need careful studies of prospective prediction of neurobehavioral outcomes to understand in whom and when to intervene. Importantly, animal models still have a role to play in preparing for gene therapies (131, 132), including the technical work to evaluate gene targeting and safety, as well as studies that restore gene expression at different points in development to understand when interventions may have an impact.
Ultimately, for any model system used to understand autism, we can only theorize about their translational potential until neurobiology-based treatments have reached the clinic. This has not yet happened in autism or in the genetic syndromes that include substantial autism risk. The closest example to date is phenylketonuria (PKU), where cellular models were sufficient to understand the impact of loss of phenylalanine hydroxylase, and where a dietary intervention was implemented that prevents cognitive impairment—and likely prevents co-occurring autism as well, based on observations in untreated individuals (133). A low-phenylalanine diet clearly leads to profound improvements in cognitive outcomes in PKU, although randomized, placebo-controlled trials were never conducted. This somewhat limits our ability to use PKU as an example of ASD treatment development.
As science continues to advance, a key question is when and how to attempt a jump across the gap from a model system to the clinic. From PKU, we might conclude that an early jump based on limited data in a model system is most feasible when the risk variant impacts a protein with a well-defined function that is relatively ubiquitous and has therefore remained consistent across species. Extrapolating from this, the cluster of autism risk genes that encode synaptic proteins is likely to be an easier set of targets than the cluster involved in chromatin or transcriptional regulation (127, 134, 135), which is likely to have a broader and harder-to-define cascade of impact. If a protein has various roles across cell types or stages of development, it is important to know that the specific roles that are targeted for translation are conserved across species, and similarities or differences in gene expression may at least be a starting point to assess this (70, 71). On the other side of the gap, the approach to testing interventions in clinical populations is also critical for making the jump successfully. Again, extrapolating from PKU, testing an intervention early in development seems likely to have a larger effect than evaluating it in adults, even though traditional drug development typically proceeds gradually from adults to adolescents to children. Objective outcome measures and predictive biomarkers are also likely to be critical, although it is difficult to have confidence about which ones will be responsive to intervention until we have treatments that work (136, 137). This challenge suggests that the first few effective interventions will not only deliver improved outcomes for specific individuals but also teach us how to better test autism treatments. This anticipated convergence of biological insight and clinical trial design provides hope that the long wait for neurobiology-based interventions will be followed by a stream of translational successes.
1. : Diagnostic and Statistical Manual of Mental Disorders, 5th ed. Washington, DC, American Psychiatric Association, 2013 Crossref, Google Scholar
2. : Autism-related language preferences of English-speaking individuals across the globe: a mixed methods investigation. Autism Res 2023; 16:406–428Crossref, Medline, Google Scholar
3. : Which terms should be used to describe autism? Perspectives from the UK autism community. Autism 2016; 20:442–462Crossref, Medline, Google Scholar
4. : Rare coding variation provides insight into the genetic architecture and phenotypic context of autism. Nat Genet 2022; 54:1320–1331Crossref, Medline, Google Scholar
5. : Prevalence and characteristics of autism spectrum disorder among children aged 8 years—Autism and Developmental Disabilities Monitoring Network, 11 sites, United States, 2018. MMWR Surveill Summ 2021; 70:1–16Crossref, Google Scholar
6. : Maternal autoantibody related autism: mechanisms and pathways. Mol Psychiatry 2019; 24:252–265Crossref, Medline, Google Scholar
7. : Maternal infection and immune involvement in autism. Trends Mol Med 2011; 17:389–394Crossref, Medline, Google Scholar
8. : Disrupted focal white matter integrity in autism spectrum disorder: a voxel-based meta-analysis of diffusion tensor imaging studies. Prog Neuropsychopharmacol Biol Psychiatry 2018; 82:242–248Crossref, Medline, Google Scholar
9. : Cortical and subcortical brain morphometry differences between patients with autism spectrum disorder and healthy individuals across the lifespan: results from the ENIGMA ASD working group. Am J Psychiatry 2018; 175:359–369Link, Google Scholar
10. : Alterations in connectome dynamics in autism spectrum disorder: a harmonized mega- and meta-analysis study using the Autism Brain Imaging Data Exchange dataset. Biol Psychiatry 2022; 91:945–955Crossref, Medline, Google Scholar
11. : Association of amygdala development with different forms of anxiety in autism spectrum disorder. Biol Psychiatry 2022; 91:977–987Crossref, Medline, Google Scholar
12. : The neuropathology of autism: a systematic review of post-mortem studies of autism and related disorders. Neurosci Biobehav Rev 2021; 129:35–62Crossref, Medline, Google Scholar
13. : Worms on the spectrum: C. elegans models in autism research. Exp Neurol 2018; 299:199–206Crossref, Medline, Google Scholar
14. : Autism spectrum disorder-related syndromes: modeling with Drosophila and rodents. Int J Mol Sci 2019; 20:4071Crossref, Medline, Google Scholar
15. : Dissecting the genetics of autism spectrum disorders: a Drosophila perspective. Front Physiol 2019; 10:987Crossref, Medline, Google Scholar
16. : A transparent window into biology: a primer on Caenorhabditis elegans. Genetics 2015; 200:387–407Crossref, Medline, Google Scholar
17. : Genetics on the fly: a primer on the Drosophila model system. Genetics 2015; 201:815–842Crossref, Medline, Google Scholar
18. : Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature 2019; 571:63–71Crossref, Medline, Google Scholar
19. : Systematic phenomics analysis of autism-associated genes reveals parallel networks underlying reversible impairments in habituation. Proc Natl Acad Sci U S A 2020; 117:656–667Crossref, Medline, Google Scholar
20. : Shank modulates postsynaptic Wnt signaling to regulate synaptic development. J Neurosci 2016; 36:5820–5832Crossref, Medline, Google Scholar
21. : A presynaptic function of shank protein in Drosophila. J Neurosci 2017; 37:11592–11604Crossref, Medline, Google Scholar
22. : Drosophila functional screening of de novo variants in autism uncovers damaging variants and facilitates discovery of rare neurodevelopmental diseases. Cell Rep 2022; 38:110517Crossref, Medline, Google Scholar
23. : Using zebrafish to model autism spectrum disorder: a comparison of ASD risk genes between zebrafish and their mammalian counterparts. Front Mol Neurosci 2020; 13:575575Crossref, Medline, Google Scholar
24. : Xenopus leads the way: frogs as a pioneering model to understand the human brain. Genesis 2021; 59:e23405Crossref, Medline, Google Scholar
25. : Learning to fish with genetics: a primer on the vertebrate model Danio rerio. Genetics 2016; 203:1069–1089Crossref, Medline, Google Scholar
26. : Zebrafish models of neurodevelopmental disorders: past, present, and future. Front Mol Neurosci 2018; 11:294Crossref, Medline, Google Scholar
27. : The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013; 496:498–503Crossref, Medline, Google Scholar
28. : Expanding the CRISPR toolbox in zebrafish for studying development and disease. Front Cell Dev Biol 2019; 7:13Crossref, Medline, Google Scholar
29. : Zebrafish as a model of neurodevelopmental disorders. Neuroscience 2020; 445:3–11Crossref, Medline, Google Scholar
30. : Zebrafish models of neurodevelopmental disorders: limitations and benefits of current tools and techniques. Int J Mol Sci 2019; 20:1296Crossref, Medline, Google Scholar
31. : Function over form: modeling groups of inherited neurological conditions in zebrafish. Front Mol Neurosci 2016; 9:55Crossref, Medline, Google Scholar
32. : Stages of embryonic development of the zebrafish. Dev Dyn 1995; 203:253–310Crossref, Medline, Google Scholar
33. : Modulation of locomotor activity in larval zebrafish during light adaptation. J Exp Biol 2007; 210:2526–2539Crossref, Medline, Google Scholar
34. : Phenotypic analysis of catastrophic childhood epilepsy genes. Commun Biol 2021; 4:680Crossref, Medline, Google Scholar
35. : Modeling autism spectrum disorders in zebrafish, in Behavioral and Neural Genetics of Zebrafish. Edited by Gerlai RT. New York, Elsevier, 2020, pp 451–480 Crossref, Google Scholar
36. : Zebrafish as a tool in the study of sleep and memory-related disorders. Curr Neuropharmacol 2022; 20:540–549Crossref, Medline, Google Scholar
37. : The gut-brain-microbiome axis and its link to autism: emerging insights and the potential of zebrafish models. Front Cel Dev Biol 2021; 9:662916Crossref, Medline, Google Scholar
38. : KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature 2012; 485:363–367Crossref, Medline, Google Scholar
39. : Estrogens suppress a behavioral phenotype in zebrafish mutants of the autism risk gene, CNTNAP2. Neuron 2016; 89:725–733Crossref, Medline, Google Scholar
40. : Pentylenetetrazole induced changes in zebrafish behavior, neural activity, and c-fos expression. Neuroscience 2005; 131:759–768Crossref, Medline, Google Scholar
41. : CRISPR/Cas9-induced shank3b mutant zebrafish display autism-like behaviors. Mol Autism 2018; 9:23Crossref, Medline, Google Scholar
42. : Disruptive CHD8 mutations define a subtype of autism early in development. Cell 2014; 158:263–276Crossref, Medline, Google Scholar
43. : Egr1 is necessary for forebrain dopaminergic signaling during social behavior. eNeuro 2022; 9Crossref, Medline, Google Scholar
44. : Phenotypic landscape of schizophrenia-associated genes defines candidates and their shared functions. Cell 2019; 177:478–491.e20Crossref, Medline, Google Scholar
45. : Rapid reverse genetic screening using CRISPR in zebrafish. Nat Methods 2015; 12:535–540Crossref, Medline, Google Scholar
46. Fmr1 knockout mice: a model to study fragile X mental retardation: the Dutch-Belgian Fragile X Consortium. Cell 1994;78:23–33Medline, Google Scholar
47. : Oxytocin receptor is not required for social attachment in prairie voles. Neuron (Online ahead of print, December 28, 2022)Google Scholar
48. : Autism-like behavioral phenotypes in BTBR T+tf/J mice. Genes Brain Behav 2008; 7:152–163Crossref, Medline, Google Scholar
49. : Social deficits, stereotypy, and early emergence of repetitive behavior in the C58/J inbred mouse strain. Behav Brain Res 2010; 208:178–188Crossref, Medline, Google Scholar
50. : Endocannabinoid modulation of orbitostriatal circuits gates habit formation. Neuron 2016; 90:1312–1324Crossref, Medline, Google Scholar
51. : Orbitofrontal and striatal circuits dynamically encode the shift between goal-directed and habitual actions. Nat Commun 2013; 4:2264Crossref, Medline, Google Scholar
52. : Social reward requires coordinated activity of nucleus accumbens oxytocin and serotonin. Nature 2013; 501:179–184Crossref, Medline, Google Scholar
53. : 5-HT release in nucleus accumbens rescues social deficits in mouse autism model. Nature 2018; 560:589–594Crossref, Medline, Google Scholar
54. : Diagnosis and management of individuals with fetal valproate spectrum disorder: a consensus statement from the European Reference Network for Congenital Malformations and Intellectual Disability. Orphanet J Rare Dis 2019; 14:180Crossref, Medline, Google Scholar
55. : Prenatal valproate in rodents as a tool to understand the neural underpinnings of social dysfunctions in autism spectrum disorder. Neuropharmacology 2019; 159:107477Crossref, Medline, Google Scholar
56. : The valproic acid-induced rodent model of autism. Exp Neurol 2018; 299:217–227Crossref, Medline, Google Scholar
57. : Maternal infection during pregnancy and risk of autism spectrum disorders: a systematic review and meta-analysis. Brain Behav Immun 2016; 58:165–172Crossref, Medline, Google Scholar
58. : Prenatal maternal infection and risk for autism in offspring: a meta-analysis. Autism Res 2021; 14:1296–1316Crossref, Medline, Google Scholar
59. : Maternal immune activation and neuropsychiatric illness: a translational research perspective. Am J Psychiatry 2018; 175:1073–1083Link, Google Scholar
60. : Maternal inflammation and neurodevelopmental programming: a review of preclinical outcomes and implications for translational psychiatry. Biol Psychiatry 2019; 85:107–121Crossref, Medline, Google Scholar
61. : Maternal immune activation causes age- and region-specific changes in brain cytokines in offspring throughout development. Brain Behav Immun 2013; 31:54–68Crossref, Medline, Google Scholar
62. : Effects of prenatal infection on brain development and behavior: a review of findings from animal models. Brain Behav Immun 2010; 24:881–897Crossref, Medline, Google Scholar
63. : The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science 2016; 351:933–939Crossref, Medline, Google Scholar
64. : Mouse models of fragile X-related disorders. Dis Model Mech 2023; 16:dmm049485Crossref, Medline, Google Scholar
65. : Mouse population genetics phenocopies heterogeneity of human Chd8 haploinsufficiency. Neuron 2023; 111:539–556.e5Crossref, Medline, Google Scholar
66. : Risperidone and aripiprazole alleviate prenatal valproic acid-induced abnormalities in behaviors and dendritic spine density in mice. Psychopharmacology (Berl) 2017; 234:3217–3228Crossref, Medline, Google Scholar
67. : Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell 2011; 147:235–246Crossref, Medline, Google Scholar
68. : Social peers rescue autism-relevant sociability deficits in adolescent mice. Autism Res 2011; 4:17–27Crossref, Medline, Google Scholar
69. : Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell 2012; 149:899–911Crossref, Medline, Google Scholar
70. : Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 2020; 583:699–710Crossref, Medline, Google Scholar
71. : Comprehensive functional genomic resource and integrative model for the human brain. Science 2018; 362:eaat8464Crossref, Medline, Google Scholar
72. : Replicable in vivo physiological and behavioral phenotypes of the Shank3B null mutant mouse model of autism. Mol Autism 2017; 8:26Crossref, Medline, Google Scholar
73. : Adult restoration of Shank3 expression rescues selective autistic-like phenotypes. Nature 2016; 530:481–484Crossref, Medline, Google Scholar
74. : Pharmacological enhancement of mGlu5 receptors rescues behavioral deficits in SHANK3 knock-out mice. Mol Psychiatry 2017; 22:784Crossref, Medline, Google Scholar
75. : Twenty years of discoveries emerging from mouse models of autism. Neurosci Biobehav Rev 2023; 146:105053Crossref, Medline, Google Scholar
76. : Of men and mice: modeling the fragile X syndrome. Front Mol Neurosci 2018; 11:41Crossref, Medline, Google Scholar
77. : Comparison of SHANK3 deficiency in animal models: phenotypes, treatment strategies, and translational implications. J Neurodev Disord 2021; 13:55Crossref, Medline, Google Scholar
78. : Current approaches and future directions for the treatment of mTORopathies. Dev Neurosci 2021; 43:143–158Crossref, Medline, Google Scholar
79. : Why primate models matter. Am J Primatol 2014; 76:801–827Crossref, Medline, Google Scholar
80. : Of mice and monkeys: using non-human primate models to bridge mouse- and human-based investigations of autism spectrum disorders. J Neurodev Disord 2012; 4:21Crossref, Medline, Google Scholar
81. : Modeling autism in non-human primates: opportunities and challenges. Autism Res 2018; 11:686–694Crossref, Medline, Google Scholar
82. : Neuroanatomy of autism. Trends Neurosci 2008; 31:137–145Crossref, Medline, Google Scholar
83. : Human prefrontal cortex: evolution, development, and pathology. Prog Brain Res 2012; 195:191–218Crossref, Medline, Google Scholar
84. : Do rats have prefrontal cortex? The Rose-Woolsey-Akert program reconsidered. J Cogn Neurosci 1995; 7:1–24Crossref, Medline, Google Scholar
85. : Evolution of prefrontal cortex. Neuropsychopharmacology 2022; 47:3–19Crossref, Medline, Google Scholar
86. : Facial expression recognition in rhesus monkeys, Macaca mulatta. Anim Behav 2009; 77:1507–1513Crossref, Medline, Google Scholar
87. : Early developmental changes in visual social engagement in infant rhesus monkeys. Dev Cogn Neurosci 2020; 43:100778Crossref, Medline, Google Scholar
88. : Altered dynamics of the fMRI response to faces in individuals with autism. J Autism Dev Disord 2016; 46:232–241Crossref, Medline, Google Scholar
89. : Face identity recognition in autism spectrum disorders: a review of behavioral studies. Neurosci Biobehav Rev 2012; 36:1060–1084Crossref, Medline, Google Scholar
90. : Stereotypic behavior in nonhuman primates as a model for the human condition. ILAR J 2014; 55:284–296Crossref, Medline, Google Scholar
91. : Nonhuman primate abnormal behavior: etiology, assessment, and treatment. Am J Primatol 2022; 84:e23380Crossref, Medline, Google Scholar
92. : The Nuremberg Code and the Nuremberg Trial: a reappraisal. JAMA 1996; 276:1662–1666Crossref, Medline, Google Scholar
93. : Frequency of prenatal loss in a macaque breeding colony. Am J Primatol 1996; 40:41–53Crossref, Medline, Google Scholar
94. : Critique of pure marmoset. Brain Behav Evol 2019; 93:92–107Crossref, Medline, Google Scholar
95. : Nonhuman primates and psychoses. J Autism Child Schizophr 1971; 1:368–375Crossref, Medline, Google Scholar
96. : Romania’s Abandoned Children: Deprivation, Brain Development, and the Struggle for Recovery. Cambridge, Mass, Harvard University Press, 2014 Crossref, Google Scholar
97. : Quasi-autistic patterns following severe early global privation. J Child Psychol Psychiatry 1999; 40:537–549Crossref, Medline, Google Scholar
98. : Naturally occurring nonhuman primate models of psychosocial processes. ILAR J 2017; 58:226–234Crossref, Medline, Google Scholar
99. : Leveraging a translational research approach to drive diagnostic and treatment advances for autism. Mol Psychiatry 2022; 27:2650–2658Crossref, Medline, Google Scholar
100. : The Macaque Social Responsiveness Scale (mSRS): a rapid screening tool for assessing variability in the social responsiveness of rhesus monkeys (Macaca mulatta). PLoS One 2016; 11:e0145956Crossref, Medline, Google Scholar
101. : A psychometrically robust screening tool to rapidly identify socially impaired monkeys in the general population. Autism Res 2020; 13:1465–1475Crossref, Medline, Google Scholar
102. : Heritability of social behavioral phenotypes and preliminary associations with autism spectrum disorder risk genes in rhesus macaques: a whole exome sequencing study. Autism Res 2022; 15:447–463Crossref, Medline, Google Scholar
103. : Reduced childhood social attention in autism model marmosets predicts impaired social skills and inflexible behavior in adulthood. Front Psychiatry 2022; 13:885433Crossref, Medline, Google Scholar
104. : Functional and molecular characterization of a non-human primate model of autism spectrum disorder shows similarity with the human disease. Nat Commun 2021; 12:5388Crossref, Medline, Google Scholar
105. : Advancing autism research from mice to marmosets: behavioral development of offspring following prenatal maternal immune activation. Front Psychiatry 2021; 12:705554Crossref, Medline, Google Scholar
106. : Activation of the maternal immune system during pregnancy alters behavioral development of rhesus monkey offspring. Biol Psychiatry 2014; 75:332–341Crossref, Medline, Google Scholar
107. : Maternal immune activation in nonhuman primates alters social attention in juvenile offspring. Biol Psychiatry 2015; 77:823–832Crossref, Medline, Google Scholar
108. : Maternal immune activation during pregnancy alters postnatal brain growth and cognitive development in nonhuman primate offspring. J Neurosci 2021; 41:9971–9987Crossref, Medline, Google Scholar
109. : Altered dendritic morphology in dorsolateral prefrontal cortex of nonhuman primates prenatally exposed to maternal immune activation. Brain Behav Immun 2023; 109:92–101Crossref, Medline, Google Scholar
110. : Maternal neuronal antibodies associated with autism and a language disorder. Ann Neurol 2003; 53:533–537Crossref, Medline, Google Scholar
111. : Autism-specific maternal autoantibodies recognize critical proteins in developing brain. Transl Psychiatry 2013; 3:e277Crossref, Medline, Google Scholar
112. : Stereotypies and hyperactivity in rhesus monkeys exposed to IgG from mothers of children with autism. Brain Behav Immun 2008; 22:806–816Crossref, Medline, Google Scholar
113. : Maternal antibodies from mothers of children with autism alter brain growth and social behavior development in the rhesus monkey. Transl Psychiatry 2013; 3:e278Crossref, Medline, Google Scholar
114. : Opportunities and limitations of genetically modified nonhuman primate models for neuroscience research. Proc Natl Acad Sci U S A 2020; 117:24022–24031Crossref, Medline, Google Scholar
115. : New models: gene-editing boom means changing landscape for primate work. Nat Med 2016; 22:1200–1202Crossref, Medline, Google Scholar
116. : Sequence diversity analyses of an improved rhesus macaque genome enhance its biomedical utility. Science 2020; 370:eabc6617Crossref, Medline, Google Scholar
117. : Modeling Rett syndrome using TALEN-edited MECP2 mutant cynomolgus monkeys. Cell 2017; 169:945–955.e10Crossref, Medline, Google Scholar
118. : Altered neurogenesis and disrupted expression of synaptic proteins in prefrontal cortex of SHANK3-deficient non-human primate. Cell Res 2017; 27:1293–1297Crossref, Medline, Google Scholar
119. : Atypical behaviour and connectivity in SHANK3-mutant macaques. Nature 2019; 570:326–331Crossref, Medline, Google Scholar
120. : Prevalence of autism spectrum disorder phenomenology in genetic disorders: a systematic review and meta-analysis. Lancet Psychiatry 2015; 2:909–916Crossref, Medline, Google Scholar
121. : FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 2015; 162:375–390Crossref, Medline, Google Scholar
122. : What have organoids and assembloids taught us about the pathophysiology of neuropsychiatric disorders? Biol Psychiatry (Online ahead of print, December 2, 2022)Google Scholar
123. : Dissecting the molecular basis of human interneuron migration in forebrain assembloids from Timothy syndrome. Cell Stem Cell 2022; 29:248–264.e7Crossref, Medline, Google Scholar
124. : Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nat Biotechnol 2020; 38:1421–1430Crossref, Medline, Google Scholar
125. : Long-term maturation of human cortical organoids matches key early postnatal transitions. Nat Neurosci 2021; 24:331–342Crossref, Medline, Google Scholar
126. : Environmental risk factors and biomarkers for autism spectrum disorder: an umbrella review of the evidence. Lancet Psychiatry 2019; 6:590–600Crossref, Medline, Google Scholar
127. : Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 2013; 155:1008–1021Crossref, Medline, Google Scholar
128. : Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 2013; 155:997–1007Crossref, Medline, Google Scholar
129. : Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson’s disease. Proc Natl Acad Sci U S A 2008; 105:5856–5861Crossref, Medline, Google Scholar
130. : Maturation and circuit integration of transplanted human cortical organoids. Nature 2022; 610:319–326Crossref, Medline, Google Scholar
131. : Adeno-associated virus (AAV)-based gene therapy products: what are toxicity studies in non-human primates showing us? Regul Toxicol Pharmacol 2023; 138:105332Crossref, Medline, Google Scholar
132. : Safety and efficacy of an engineered hepatotropic AAV gene therapy for ornithine transcarbamylase deficiency in cynomolgus monkeys. Mol Ther Methods Clin Dev 2021; 23:135–146Crossref, Medline, Google Scholar
133. : Inborn errors of metabolism associated with autism spectrum disorders: approaches to intervention. Front Neurosci 2021; 15:673600Crossref, Medline, Google Scholar
134. : Synaptic, transcriptional, and chromatin genes disrupted in autism. Nature 2014; 515:209–215Crossref, Medline, Google Scholar
135. : Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron 2011; 70:898–907Crossref, Medline, Google Scholar
136. : In search of biomarkers to guide interventions in autism spectrum disorder: a systematic review. Am J Psychiatry 2023; 180:23–40Link, Google Scholar
137. : The Autism Biomarkers Consortium for Clinical Trials: initial evaluation of a battery of candidate EEG biomarkers. Am J Psychiatry 2023; 180:41–49Link, Google Scholar



