Epigenetic Aging in Major Depressive Disorder
Abstract
Objective:
Major depressive disorder is associated with an increased risk of mortality and aging-related diseases. The authors examined whether major depression is associated with higher epigenetic aging in blood as measured by DNA methylation (DNAm) patterns, whether clinical characteristics of major depression have a further impact on these patterns, and whether the findings replicate in brain tissue.
Method:
DNAm age was estimated using all methylation sites in blood of 811 depressed patients and 319 control subjects with no lifetime psychiatric disorders and low depressive symptoms from the Netherlands Study of Depression and Anxiety. The residuals of the DNAm age estimates regressed on chronological age were calculated to indicate epigenetic aging. Major depression diagnosis and clinical characteristics were assessed with questionnaires and psychiatric interviews. Analyses were adjusted for sociodemographic characteristics, lifestyle, and health status. Postmortem brain samples of 74 depressed patients and 64 control subjects were used for replication. Pathway enrichment analysis was conducted using ConsensusPathDB to gain insight into the biological processes underlying epigenetic aging in blood and brain.
Results:
Significantly higher epigenetic aging was observed in patients with major depression compared with control subjects (Cohen’s d=0.18), with a significant dose effect with increasing symptom severity in the overall sample. In the depression group, epigenetic aging was positively and significantly associated with childhood trauma score. The case-control difference was replicated in an independent data set of postmortem brain samples. The top significantly enriched Gene Ontology terms included neuronal processes.
Conclusions:
As compared with control subjects, patients with major depression exhibited higher epigenetic aging in blood and brain tissue, suggesting that they are biologically older than their corresponding chronological age. This effect was even more profound in the presence of childhood trauma.
A growing body of literature suggests that major depressive disorder is associated with increased risks of mortality and aging-related phenotypes and diseases, including cardiovascular disease, diabetes, obesity (1), cancer (2), cognitive impairment (3), and frailty (4). Given the associated negative impact on quality of life and health care costs (5), it is of interest to investigate whether patients with major depression are prone to accelerated aging.
The literature provides evidence for advanced biological aging in major depressive disorder, as indicated by shorter telomere length (6, 7) and advanced brain aging (8). Recently, alternative markers of biological age derived from DNA methylation (also known as “epigenetic clocks”) have been developed. Chronological age can be accurately predicted from methylation data and yields estimates of DNA methylation age (DNAm age) (9, 10). Furthermore, DNAm age can be studied as either “decelerated” or “accelerated” by regressing it on chronological age to obtain a measure of epigenetic aging. Thus, DNAm age is a promising candidate for reliably investigating accelerated or premature aging in major depression.
Previous studies have shown epigenetic aging in individuals with Down’s syndrome (11), HIV-positive patients (12), and individuals with obesity (13). In addition, epigenetic aging has been associated with poorer physical and cognitive fitness (14), increased smoking and alcohol use (15), cancer (16), Alzheimer’s disease (17), cardiovascular disease (16), and an increased risk of mortality (18). A few studies have investigated DNAm age in relation to schizophrenia (19, 20), life stress (21), and posttraumatic stress disorder (22, 23), with mixed findings. However, studies examining epigenetic aging in relation to major depressive disorder are currently lacking.
In this study, we examined whether major depression is associated with higher epigenetic aging in blood, using a large, clinically well phenotyped sample to further explore associations with clinical characteristics, and we sought to replicate the findings in postmortem brain tissue. We used a sequencing-based approach (24, 25) that yields almost complete coverage of the CpG methylome, which allowed us to obtain the most accurate DNAm age estimates for our sample and to better explore the biological processes underlying epigenetic aging.
Method
The Netherlands Study of Depression and Anxiety
Study participants were from the Netherlands Study of Depression and Anxiety (NESDA), an ongoing longitudinal multicenter cohort study designed to investigate the long-term course and consequences of depressive and anxiety disorders (26). NESDA’s 2,981 participants (ages 18–65) include patients with a current or lifetime diagnosis of depression and/or an anxiety disorder and control subjects with no lifetime depressive disorder or anxiety disorders. Participants were recruited from the general population, general practices, and mental health organizations in order to reflect various settings and the entire range of psychopathology. Presence of major depression was ascertained with the DSM-IV-based Composite International Diagnostic Interview, version 2.1 (CIDI) (27), administered by trained research staff. Exclusion criteria were a clinically overt primary diagnosis of other psychiatric conditions (e.g., psychotic disorders, obsessive-compulsive disorder, bipolar disorder, and severe substance use disorders) and not being fluent in Dutch. The study was approved by the ethical committees of all participating centers, and participants provided written informed consent.
The total sample of 1,130 participants was divided into a control group (no lifetime psychiatric disorders and low depressive symptoms, as indicated by a score <14 on the Inventory of Depressive Symptomatology [28]) (N=319) and a group with current major depressive disorder (a score ≥14 on the Inventory of Depressive Symptomatology within the past 6 months) (N=811). Individuals who did not meet criteria for either of the two groups were excluded. The sample selection was further based on good-quality genome-wide association study genotype information available from a previous investigation (29).
Assessed Phenotypes
Data on sex, education level (in years), and body mass index (BMI) were collected during interviews. Alcohol use was recorded as mean number of drinks per week. Smoking behavior was measured by plasma cotinine levels, an adequate marker for calculating recent tobacco exposure (30). Physical activity was assessed using the International Physical Activity Questionnaire and indicated by total metabolic equivalent (MET) minutes per week. Health status was assessed as number of chronic diseases for which participants received medical treatment.
In all participants, depression severity was measured with the self-report version of the 30-item Inventory of Depressive Symptomatology (28). Childhood trauma was assessed using the childhood trauma interview from the Netherlands Mental Health Survey and Incidence Study, with personal history questions that included a structured inventory of trauma exposure during childhood. Finally, use of antidepressants was assessed through container inspection and categorized using the World Health Organization’s Anatomical Therapeutic Chemical Classification System: tricyclic antidepressants, selective serotonin reuptake inhibitors, and other antidepressants.
In participants with major depressive disorder, the duration of depression was measured by the Life Chart Interview, utilizing a calendar method to assess the percentage of time in which symptoms were present during the past 4 years (31). Current comorbid anxiety (panic disorder, generalized anxiety disorder, agoraphobia, social phobia) and alcohol disorder, as well as age at onset of depression were assessed with the CIDI. A more detailed description of all phenotypes is provided in the data supplement that accompanies the online edition of this article.
DNA Methylation Measurements
To assay the methylation status of the approximately 28 million common CpG sites in the human genome, we used an optimized protocol for MBD-seq (25). With this approach, genomic DNA is fragmented and the methylated fragments are then bound to the MBD2 protein, which has a high affinity for methylated DNA. The nonmethylated fraction is washed away, and only the methylation-enriched fraction is sequenced (for more details, see the online data supplement). This optimized protocol assesses about 94% of the CpGs in the methylome (25). The sequenced reads were aligned to the reference genome (build hg19/GRCh37) with Bowtie 2 (32) using local and gapped alignment. Aligned reads were further processed using the RaMWAS Bioconductor package (33) to perform quality control and calculate methylation scores for each CpG.
DNAm Age Estimation
While existing algorithms (9, 10) have gone through demonstrations of utility and reliability, estimating DNAm age with those prediction models would have been suboptimal for this study. These algorithms were derived using methylation data from a different platform in study populations with different characteristics (e.g., age distribution). Commonly used methods for assaying DNA methylation depend on the Illumina arrays, platforms that generate variables representing percentage methylated (ranging from 0 to 1). We used MBD-seq, generating methylation data that is semiquantitative (scores may range from 0–20) (24). Because the weights assigned to individual CpGs when making age predictions directly depend on the platform and study population, they will not optimally capture the effects of CpGs on age in the present study. Therefore, we “recalibrated” the DNAm age estimate in a way that is optimal for this study. It is important to emphasize that we aimed to obtain the best possible DNAm age estimates for MBD-seq data in our sample. We have not developed a new clock to be generalized to data from platforms other than MBD-seq. Furthermore, because we already collected MBD-seq data, we did not attempt to reduce the predictor set to the smallest number of CpGs, as pruning sites may reduce the precision of the DNAm age estimates.
Our approach for estimating DNAm age is similar to the one taken by Horvath (10). Specifically, we used elastic nets, a variable selection method that is particularly useful when the number of predictors is much larger than the number of observations (34). Parameter alpha was set to zero (i.e., ridge regression, retaining all sites in the model) where chronological age was used as outcome and methylation sites as predictors. To estimate predictive power and obtain DNAm age estimates for each subject, k-fold cross-validation was used, with k=10. Thus, the sample was randomly partitioned into 10 equally sized subsamples. Of the 10 subsamples, nine were used as training data and the remaining subsample as validation data. This ensured that in samples with the same properties and platform, our results would “replicate” and provide unbiased estimates of DNAm age. In the RaMWAS implementation, a cycle of methylome-wide association studies (MWAS), marker selection, and estimation via ridge regression was repeated in each training data set, with the resulting model applied to the test data to obtain unbiased estimates of DNAm age for each of the k=10 iterations.
Validation of the Use of MBD-seq Data to Estimate DNAm Age
Several analyses were performed to validate the model. First, the model used to estimate DNAm age contained 80,000 CpGs (see Table S1 in the data supplement). Tenfold cross-validation showed that chronological age could be predicted very well, with a correlation of 0.95 (p<0.001) (Figure 1). Second, when analyzing assessed phenotypes in NESDA with DNAm age, we confirmed some similar determinants of DNAm age found in previous studies validating our outcome measure (see Table S3 in the data supplement). Male sex (35) and higher BMI (13, 36, 37) were associated with higher epigenetic aging. Third, to validate calculation of DNAm age, we used both ridge regression and the lasso method (used by Horvath). The additional elastic net model, with parameter alpha set to 0.5, resulted in a comparable correlation of 0.93 between chronological age and predicted DNAm age in our data set (compared with 0.95 with ridge regression and alpha=0), indicating that parameter set point did not have a large impact on our outcome measure. Finally, to ensure that no systematic bias was introduced by training the model on both case subjects and control subjects, we also trained the prediction model in control subjects only. This resulted in a slightly lower correlation between DNAm age estimates and chronological age (r=0.93), since the control subjects represented only a third of our total sample (i.e., lower statistical power). However, the correlation between the DNAm age estimates obtained in the full sample and those obtained using only control subjects was high (r=0.98, p<0.001), indicating that psychiatric status did not have an impact on the estimation of DNAm age.
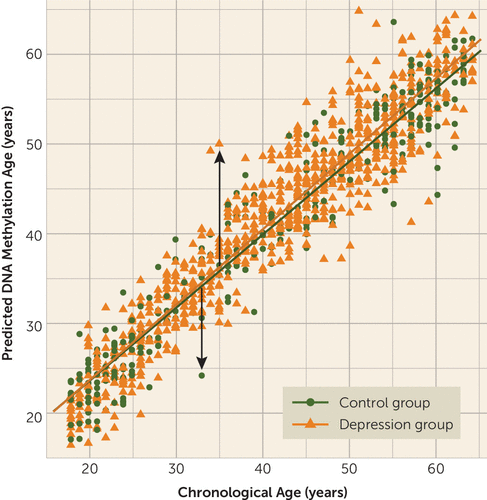
FIGURE 1. DNA Methylation Age Prediction Using Methyl-CpG Binding Domain Protein-Enriched Genome Sequencing (MBD-seq) in the Netherlands Study of Depression and Anxietya
a The plot shows the prediction of DNA methylation (DNAm) age using MBD-seq across groups in blood. Each circle or triangle represents an individual subject (N=1,130), and the lines indicate regression lines (control group [N=319]: r=0.94, p<0.001; major depression group [N=811]: r=0.96, p<0.001). The arrows indicate the outcome variable epigenetic aging, representing higher epigenetic aging if the individual’s estimated DNAm age exceeds chronological age (upward arrow), whereas negative epigenetic aging indicates lower epigenetic aging (downward arrow).
Postmortem Brain Samples
We pooled data from five brain sample collections from four different brain banks (the Victorian Brain Bank Network, the Harvard Brain Bank, the Netherlands Brain Bank, and the Stanley Medical Research Institute), including a total of 141 brain samples from the Brodmann’s areas 10 and 25 regions. Presence of major depressive disorder (N=74) was determined by at least one psychiatrist by using information obtained from a family member who was well acquainted with the deceased. Control subjects (N=67) had no history of psychiatric disorders. Postmortem interval and pH were recorded in the brain collections, with the exception of the Harvard Brain Bank.
To further test the reliability and validity of our methods, we used the same approach to predict DNAm age in these samples with MBD-seq methylation data generated from the SOLiD 5500 W platform (Life Technologies, Carlsbad, Calif.). The model used to predict DNAm age contained 100,000 CpGs (see Table S2 in the data supplement) and obtained a correlation of 0.69 between predicted DNAm age and chronological age. The lower correlation with age is likely because the methylation data were generated with an older platform with lower quality (e.g., lower alignment of reads) from a more heterogeneous data set. More details about the samples and methods are provided in the data supplement.
Statistical Analyses for Discovery
To investigate case-control differences in epigenetic aging, we conducted linear regression models with epigenetic aging as the outcome and all covariates as predictors. To correct for the relative abundance of cell types that may be differentially associated with major depression, an additional model included cell type proportions as covariates (38). Other linear regression models were used to examine the relationship between epigenetic aging and Inventory of Depressive Symptomatology score across groups and clinical characteristics within patients with major depression. All analyses were corrected for all sociodemographic, lifestyle, and health covariates and used two-tailed tests with a significance threshold of 0.05.
Statistical Analyses for Replication
Within postmortem brain samples, we constructed a linear mixed model in R using the nlme package to account for the heterogeneity of epigenetic aging across brain collections. Thus, brain collection was entered as random effect and sex as fixed effect. The p value was derived by a likelihood ratio test, a hypothesis-driven one-sided test, with p<0.05 considered significant.
Bioinformatics Analyses
To perform enrichment tests of top MWAS findings in brain and blood, we used the shiftR R package with 1 million permutations for each test and used three thresholds (0.5%, 1%, and 5%) to define “top findings.” To account for this “multiple testing,” shiftR uses the same thresholds in the permutations where the test statistic distribution under the null hypothesis is generated from the most significant thresholds or combination of thresholds. A more in-depth description is provided in the data supplement. To gain insight into the overlapping biological pathways affecting epigenetic aging in blood and brain, we used ConsensusPathDB (39) to test whether genes harboring epigenetic aging–associated CpGs were enriched for level 5 Gene Ontology (GO) terms. Methylation sites with p<1×10−5 were selected and had to be within gene boundaries. At least four genes had to be present in the GO term to be considered. Finally, we also evaluated the overlap between chronological age–associated and epigenetic aging–associated CpG sites to examine whether similar biological processes were involved in chronological and biological aging.
Results
Higher Epigenetic Aging and Major Depression in NESDA
The mean age of the NESDA sample was 41.5 years (SD=13.0, range=18–64), and 64.5% of the sample were female (Table 1). The depression and control groups did not differ significantly in age, but the depression group had a higher proportion of females (p=0.02) and had fewer years of education on average (p<0.001). As anticipated, patients in the depression group reported higher levels of depression severity and use of antidepressants (all p values, <0.001). The mean childhood trauma score was also higher in the depressed group (p<0.001).
| Characteristic | Control Group (N=319) | Depression Group (N=811) | ||
|---|---|---|---|---|
| Sociodemographic characteristics | ||||
| N | % | N | % | |
| Female | 188 | 58.9 | 541 | 66.7 |
| Mean | SD | Mean | SD | |
| Age (years) | 41.6 | 14.63 | 41.5 | 12.26 |
| Education level (years) | 13.1 | 3.15 | 11.5 | 3.20 |
| Mean | SD | Mean | SD | |
| Lifestyle and health characteristics | ||||
| Body mass index | 25.2 | 4.50 | 25.9 | 5.31 |
| Cotinine levels (ng/mL) | 70.9 | 200.8 | 103.3 | 183.9 |
| Alcohol intake (mean number of drinks per week) | 7.10 | 7.11 | 6.32 | 9.12 |
| Physical activity (1,000 MET minutes per week) | 3.91 | 2.86 | 3.49 | 3.17 |
| Number of chronic diseases | 0.46 | 0.74 | 0.69 | 0.92 |
| Mean | SD | Mean | SD | |
| Clinical characteristics | ||||
| Inventory of Depressive Symptomatology score | 5.02 | 3.54 | 33.8 | 10.9 |
| Childhood trauma score | 0.30 | 0.69 | 1.23 | 1.24 |
| Age at depression onset (years) | 27.0 | 12.5 | ||
| Symptom duration (% of time in past 4 years) | 0.39 | 0.30 | ||
| N | % | N | % | |
| Comorbid anxiety disorder | 538 | 66.4 | ||
| Comorbid alcohol disorder | 270 | 33.3 | ||
| Antidepressant use | ||||
| Tricyclic antidepressants | 0 | 0.0 | 38 | 4.7 |
| Selective serotonin reuptake inhibitors | 1 | 0.3 | 243 | 30.0 |
| Other antidepressants | 0 | 0.0 | 90 | 11.2 |
| Mean | SD | Mean | SD | |
| Epigenetic aging | –0.45 | 3.37 | 0.18 | 3.65 |
TABLE 1. Characteristics of Participants With and Without Depression in the Netherlands Study of Depression and Anxiety in an Analysis of Epigenetic Aging in Major Depressive Disordera
Epigenetic aging showed (by design) a mean of zero (SD=3.58 years), ranging from −13.26 to 15.00. Patients in the depression group had significantly higher epigenetic aging compared with the control group (b=0.64, t=2.65, p=0.008; effect size, Cohen’s d=0.18), indicating that the patient was estimated to be 0.64 years (or 7.68 months) older on average than the control group after full adjustment for covariates (Table 2). Additional analyses correcting for cell type proportions did not change results and produced a Cohen’s d of 0.14 (see the data supplement). Consistent with a dose-response effect, a fully adjusted linear regression showed that greater epigenetic aging was significantly associated with higher Inventory of Depressive Symptomatology score in the overall sample (β=0.10, p=0.001). As expected from the high correlation between the DNAm age estimates generated by both models (r=0.98), the above-mentioned results remained unchanged when the same analyses were performed with the DNAm age estimates from the control-subjects-only model (see the data supplement).
| Control Group (N=319) | Depression Group (N=811) | Depression Versus Control Group | Inventory of Depressive Symptomatology Score | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Model | Mean | SE | 95% CI | Mean | SE | 95% CI | p | Cohen’s d | β | p |
| Basic adjusteda | –0.51 | 0.20 | –0.90, –0.11 | 0.20 | 0.13 | –0.05, 0.45 | 0.004 | 0.20 | 0.12 | <0.001 |
| Fully adjustedb | –0.46 | 0.20 | –0.86, –0.06 | 0.18 | 0.13 | –0.07, 0.43 | 0.008 | 0.18 | 0.10 | 0.001 |
TABLE 2. Estimated Marginal Means of Epigenetic Aging, by Major Depressive Disorder Status and Association With Depression Severity in the Overall Sample, in Basic and Fully Adjusted Analyses
Exploratory Analyses of Epigenetic Aging and Clinical Characteristics
Within the depression groups, we found epigenetic aging to be positively associated with childhood trauma score (β=0.09, p=0.02, see Table 3). The association between epigenetic aging and Inventory of Depressive Symptomatology score in the overall sample did not remain significant when analyzed only within the depression group (β=0.05, p=0.21), likely because of lower variation in symptom severity. No further significant associations with clinical characteristics were found.
| Variable | β | pb |
|---|---|---|
| Inventory of Depressive Symptomatology score | 0.05 | 0.21 |
| Duration of illness | –0.02 | 0.58 |
| Age at onset | 0.03 | 0.42 |
| Comorbid anxiety disorder | –0.02 | 0.53 |
| Comorbid alcohol dependence disorder | 0.05 | 0.21 |
| Childhood trauma score | 0.09 | 0.02 |
| Antidepressant use | ||
| Tricyclic antidepressants | 0.02 | 0.67 |
| Selective serotonin reuptake inhibitors | –0.04 | 0.31 |
| Other antidepressants | –0.04 | 0.29 |
TABLE 3. Associations Between Epigenetic Aging and Clinical Characteristics in Patients With Major Depressive Disorder (N=811)a
Further analyses revealed that patients in the depression group who had childhood trauma showed the highest epigenetic aging compared with control subjects without childhood trauma (p=0.001, Cohen’s d=0.29), highlighting the fact that this major depression and childhood trauma subgroup is associated with the highest epigenetic aging (see Figure S1 in the data supplement). Notably, greater severity of symptoms of chronic major depression was correlated with childhood trauma (r=0.39, p<0.001), making it difficult to discern which of these two factors drives increased epigenetic aging. Linear regression indeed showed that both childhood trauma (β=0.08, p=0.01) and depression severity score (β=0.07, p=0.03) were significant predictors of epigenetic aging when analyzed in the same model.
Replication in Postmortem Brain Samples
The mean age of the postmortem brain samples was 55.2 years (SD=19.3, range=20–100), and 45.4% of the sample were female. Groups were matched on age and sex. The mean postmortem interval was 35.1 hours (SD=21.1), and the mean pH was 6.51 (SD=0.25) across samples. Epigenetic aging was uncorrelated with pH or postmortem interval. Table 4 summarizes the descriptive characteristics by brain collection. Only the control (N=67) and depression (N=74) samples from the same brain collection were included in analyses (see the data supplement for more detail).
| Brain Collection and Characteristic | Control Group (N=67) | Depression Group (N=74) | ||
|---|---|---|---|---|
| Brain collection 1b | N=30 | N=30 | ||
| Mean | SD | Mean | SD | |
| Age (years) | 51.63 | 12.94 | 51.93 | 18.61 |
| Postmortem interval (hours) | 47.04 | 14.63 | 41.74 | 15.74 |
| pH | 6.32 | 0.21 | 6.50 | 0.28 |
| N | % | N | % | |
| Female | 17 | 56.7 | 18 | 60.0 |
| Brain collection 2 | N=4 | N=3 | ||
| Mean | SD | Mean | SD | |
| Age (years) | 77.00 | 12.25 | 83.00 | 13.00 |
| Postmortem interval (hours) | NA | NA | NA | NA |
| pH | NA | NA | NA | NA |
| N | % | N | % | |
| Female | 1 | 25.0 | 1 | 33.3 |
| Brain collection 3 | N=9 | N=9 | ||
| Mean | SD | Mean | SD | |
| Age (years) | 85.67 | 8.57 | 86.44 | 8.37 |
| Postmortem interval (hours) | 5.28 | 0.78 | 6.59 | 1.93 |
| pH | 6.56 | 0.15 | 6.49 | 0.23 |
| N | % | N | % | |
| Female | 6 | 66.7 | 2 | 66.7 |
| Brain collection 4 | N=11 | N=22 | ||
| Mean | SD | Mean | SD | |
| Age (years) | 48.00 | 11.95 | 42.32 | 11.15 |
| Postmortem interval (hours) | 26.72 | 9.79 | 30.14 | 12.84 |
| pH | 6.64 | 0.19 | 6.65 | 0.13 |
| N | % | N | % | |
| Female | 4 | 36.4 | 10 | 45.5 |
| Brain collection 5 | N=13 | N=10 | ||
| Mean | SD | Mean | SD | |
| Age (years) | 51.15 | 8.35 | 48.80 | 8.35 |
| Postmortem interval (hours) | 24.00 | 15.00 | 49.00 | 49.50 |
| pH | 6.69 | 0.18 | 6.64 | 0.23 |
| N | % | N | % | |
| Female | 0 | 0.0 | 1 | 10.0 |
TABLE 4. Descriptive Characteristics of the Postmortem Brain Samples in an Analysis of Epigenetic Aging in Major Depressive Disordera
Our replication findings in independent brain samples supported our findings in NESDA and again showed that the epigenetic aging was higher in the depression group than in the control group (b=1.11, χ2=3.41, p=0.03). The beta indicates that the depression group was estimated to be on average 1.11 years older than the control group. The phenotype information available from the postmortem samples was limited, and therefore we were unable to attempt any replication of the exploratory clinical associations observed, such as childhood trauma, in NESDA.
Enrichment Testing and Gene Ontology Analyses
When evaluating the overlap between both epigenetic aging indicators, we found that after correcting for multiple testing, the top 1% findings from the epigenetic aging MWAS in blood were significantly enriched for CpGs in the top 0.5% of the epigenetic aging MWAS from brain (odds ratio=1.19, p<0.001). To examine possible processes underlying epigenetic aging in both tissues, we performed pathway analyses on the 1,084 overlapping CpGs associated with epigenetic aging, leading to 330 genes (90.7%) that were present in at least one GO category. Subsequently, this resulted in 53 significantly enriched GO terms (see Table S4 in the data supplement). The top GO terms included neurogenesis (p=9.79×10−9), neuron differentiation (p=5.34×10−8), and regulation of neuron death (p=4.67×10−5), indicating that several depression-relevant pathways were enriched in the cross-tissue epigenetic aging indicators.
Discussion
To the best of our knowledge, this is the first time higher epigenetic aging in patients with major depression compared with control subjects has been demonstrated. Exploratory analyses suggested even more pronounced epigenetic aging in patients with major depression who had higher childhood trauma scores. The case-control difference in blood was replicated in postmortem brain tissue. Finally, analyses showed significantly enriched neuronal pathways associated with the overlap between epigenetic aging–associated CpGs from blood and brain tissue.
Replication of our main finding in postmortem brain tissue bolstered confidence in the observed higher epigenetic aging in major depression. Moreover, the significantly enriched overlap suggests that at least some processes affecting epigenetic aging are at play in both blood and brain. There is some evidence that blood and brain show concordance in methylation (40) and epigenetic aging (10). However, considering the interactions between stress, central and peripheral immune processes, and neurobiology (41), it is plausible and likely that epigenetic aging in major depression is also dictated by many systemic processes. Nonetheless, more work is needed to confirm the higher epigenetic aging findings and to better characterize advanced aging–associated genes and their implications in major depression.
DNAm age is just one of the several available markers of biological aging (35). The present study confirms advanced or premature biological aging in major depression with a novel platform and is consistent with the literature regarding telomere length as biological marker of aging in major depression (7, 42). Also in line with other studies (43, 44), post hoc analyses between telomere length and epigenetic aging showed nonsignificant relationships, suggesting that both measures likely independently track different aspects of biological aging. Similarly, other post hoc analyses showed that telomere length did not alter our findings when accounted for, providing further evidence that epigenetic aging captures significant aging signal different from telomere length (see the data supplement).
We found that childhood trauma was positively associated with higher epigenetic aging in patients with major depression. It seems conceivable that major depression and accumulated stress throughout the lifetime due to childhood trauma may alter the epigenetic landscape and influence genomic regulation and function (45). However, this study did not identify additional relationships between higher epigenetic aging and more cumulative clinical characteristics, such as earlier onset age or longer duration of depression. Rather, our findings suggest that higher epigenetic aging in major depression may be driven largely by severity of illness.
Alternatively, childhood trauma may produce long-lasting epigenetic “scars” that have an impact on major depression and advanced or premature aging processes later in life. Individuals with childhood trauma and depressive disorders have an earlier onset age, higher symptom severity, more comorbidities, increased suicidality, and poorer treatment response than patients without childhood trauma (46, 47). As Teicher and Samson (47) have suggested, presence of childhood trauma is associated with a clinically and neurobiologically distinct subtype of depression.
Strikingly, three out of 10 top GO categories enriched across tissues included neuronal pathways. Epigenetic mechanisms are critical in early brain development, adult neurogenesis, and late-stage brain maturation (48). Given that these are all processes that seem markedly aberrant in major depression (49), the implicated pathways suggest that epigenetic aging in major depression directly contributes to symptomology. Additionally, the degree of overlap between the top 1% findings of epigenetic aging from blood and brain and the top 0.5% findings of chronological age in NESDA (odds ratio=85.31, p<0.001, and odds ratio=1.64, p<0.001) was highly significant, suggesting that biological aging overlaps with the same epigenetic processes that underlie chronological aging.
Although effect sizes such as those we observed here are common in depression research (e.g., oxidative stress, brain-derived neurotrophic factor, and cortisol yield effect sizes ranging from 0.15 to 0.31 [50–52]), it is possible that this is an underestimate. The reason is that DNAm age is estimated from the residuals of the regression of methylation data on chronological age. This residual variance comprises two components: true unique variance associated with DNAm age, and measurement errors. However, because the residual variance is very small (the correlation between chronological age and methylation data was 0.95), even small measurement errors can have a large negative effect on the reliability of the DNAm age that is defined as reliability=Var(DNAm age)/(Var[DNAm age]+Var[errors of measurement]). As a less than perfect reliability will attenuate the correlation of DNAm age and depression status, the real effect size may have been underestimated.
Strengths of this study are the replication in postmortem brain tissue and inclusion of a large, clinically well characterized, and representative sample, with data that included many potential confounders that did not explain our findings. Given the full methylome coverage, we were also able to examine which biological pathways seemingly underlie epigenetic aging. However, our findings should also be considered against some limitations. A direct comparison of our MBD-seq-based epigenetic clock and existing Illumina array-based clocks was not possible and was beyond the scope of this study. However, a side-by-side comparison is an interesting endeavor for a future methodological study. Furthermore, with the present cross-sectional data, we were not able to disentangle whether greater epigenetic aging in major depression truly reflects aging acceleration over time or whether some individuals have increased epigenetic aging from birth or before adulthood that continues to be stable thereafter (53). Further studies with longitudinal designs are needed to distinguish the two possibilities.
In conclusion, our findings show that DNAm age from both blood and brain of patients with major depression is higher than their corresponding chronological age, which may contribute to their increased risk for mortality and aging-related diseases. Furthermore, higher childhood trauma scores correlated with higher epigenetic aging in patients with major depression. Taken together, our findings suggest that higher methylation aging in major depression is present in both blood and brain and that higher epigenetic aging largely overlaps with the same underpinnings associated with chronological aging. Further research is needed to investigate the causal relationships between age-associated alterations in DNA methylation and major depression.
1 : Depression and cardiovascular disease: epidemiological evidence on their linking mechanisms. Neurosci Biobehav Rev 2017; 74(Pt B):277–286Crossref, Medline, Google Scholar
2 : Depression and cancer: mechanisms and disease progression. Biol Psychiatry 2003; 54:269–282Crossref, Medline, Google Scholar
3 : A meta-analysis of depression severity and cognitive function. J Affect Disord 2009; 119:1–8Crossref, Medline, Google Scholar
4 : The relationship between depression and frailty syndrome: a systematic review. Aging Ment Health 2015; 19:762–772Crossref, Medline, Google Scholar
5 : Epigenetic genome-wide association methylation in aging and longevity. Epigenomics 2012; 4:503–509Crossref, Medline, Google Scholar
6 : The association between depression and leukocyte telomere length: a meta-analysis. Depress Anxiety 2015; 32:229–238Crossref, Medline, Google Scholar
7 : Depression and telomere length: a meta-analysis. J Affect Disord 2016; 191:237–247Crossref, Medline, Google Scholar
8 : Accelerated brain aging in schizophrenia and beyond: a neuroanatomical marker of psychiatric disorders. Schizophr Bull 2014; 40:1140–1153Crossref, Medline, Google Scholar
9 : Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 2013; 49:359–367Crossref, Medline, Google Scholar
10 : DNA methylation age of human tissues and cell types. Genome Biol 2013; 14:R115Crossref, Medline, Google Scholar
11 : Accelerated epigenetic aging in Down syndrome. Aging Cell 2015; 14:491–495Crossref, Medline, Google Scholar
12 : Accelerated epigenetic aging in brain is associated with pre-mortem HIV-associated neurocognitive disorders. J Neurovirol 2015; 22:366–375Crossref, Medline, Google Scholar
13 : Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci USA 2014; 111:15538–15543Crossref, Medline, Google Scholar
14 : The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int J Epidemiol 2015; 44:1388–1396Crossref, Medline, Google Scholar
15 : Methylomic aging as a window onto the influence of lifestyle: tobacco and alcohol use alter the rate of biological aging. J Am Geriatr Soc 2015; 63:2519–2525Crossref, Medline, Google Scholar
16 : Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin Epigenetics 2016; 8:64Crossref, Medline, Google Scholar
17 : Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer’s disease related cognitive functioning. Aging (Albany NY) 2015; 7:1198–1211Crossref, Medline, Google Scholar
18 : DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY) 2016; 8:1844–1865Crossref, Medline, Google Scholar
19 : Epigenetic analysis confirms no accelerated brain aging in schizophrenia. NPJ Schizophr 2017; 3:26Crossref, Medline, Google Scholar
20 : DNA methylation age is not accelerated in brain or blood of subjects with schizophrenia. Schizophr Res (Epub ahead of print, Oct 5, 2017)Google Scholar
21 : Lifetime stress accelerates epigenetic aging in an urban, African American cohort: relevance of glucocorticoid signaling. Genome Biol 2015; 16:266Crossref, Medline, Google Scholar
22 : Accelerated DNA methylation age: associations with PTSD and neural integrity. Psychoneuroendocrinology 2016; 63:155–162Crossref, Medline, Google Scholar
23 : Longitudinal changes of telomere length and epigenetic age related to traumatic stress and post-traumatic stress disorder. Psychoneuroendocrinology 2015; 51:506–512Crossref, Medline, Google Scholar
24 : MBD-seq as a cost-effective approach for methylome-wide association studies: demonstration in 1500 case-control samples. Epigenomics 2012; 4:605–621Crossref, Medline, Google Scholar
25 : Enrichment methods provide a feasible approach to comprehensive and adequately powered investigations of the brain methylome. Nucleic Acids Res 2017; 45:e97Crossref, Medline, Google Scholar
26 : The Netherlands Study of Depression and Anxiety (NESDA): rationale, objectives, and methods. Int J Methods Psychiatr Res 2008; 17:121–140Crossref, Medline, Google Scholar
27 : Reliability and validity studies of the WHO–Composite International Diagnostic Interview (CIDI): a critical review. J Psychiatr Res 1994; 28:57–84Crossref, Medline, Google Scholar
28 : The Inventory of Depressive Symptomatology (IDS): psychometric properties. Psychol Med 1996; 26:477–486Crossref, Medline, Google Scholar
29 : Genome-wide association of major depression: description of samples for the GAIN Major Depressive Disorder Study: NTR and NESDA biobank projects. Eur J Hum Genet 2008; 16:335–342Crossref, Medline, Google Scholar
30 : Optimal serum cotinine levels for distinguishing cigarette smokers and nonsmokers within different racial/ethnic groups in the United States between 1999 and 2004. Am J Epidemiol 2009; 169:236–248Crossref, Medline, Google Scholar
31 : The Life Chart Interview: a standardized method to describe the course of psychopathology. Int J Methods Psychiatr Res 1994; 4:143–155Google Scholar
32 : Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9:357–359Crossref, Medline, Google Scholar
33 : RaMWAS: fast methylome-wide association study pipeline for enrichment platforms. Bioinformatics (Epub ahead of print, Feb 12, 2018)Google Scholar
34 : Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010; 33:1–22Crossref, Medline, Google Scholar
35 : Biological age predictors. EBioMedicine 2017; 21:29–36Crossref, Medline, Google Scholar
36 : Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany NY) 2017; 9:419–446Crossref, Medline, Google Scholar
37 : Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol 2014; 15:R24Crossref, Medline, Google Scholar
38 : Correcting for cell-type effects in DNA methylation studies: reference-based method outperforms latent variable approaches in empirical studies. Genome Biol 2017; 18:24Crossref, Medline, Google Scholar
39 : The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res 2013; 41:D793–D800Crossref, Medline, Google Scholar
40 : On the outside, looking in: a review and evaluation of the comparability of blood and brain “-omes.” Am J Med Genet B Neuropsychiatr Genet 2013; 162:595–603Crossref, Google Scholar
41 : Integrating neuroimmune systems in the neurobiology of depression. Nat Rev Neurosci 2016; 17:497–511Crossref, Medline, Google Scholar
42 : Peripheral telomere length and hippocampal volume in adolescents with major depressive disorder. Transl Psychiatry 2015; 5:e676Crossref, Medline, Google Scholar
43 : Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clin Epigenetics 2016; 8:21Crossref, Medline, Google Scholar
44 : The epigenetic clock and telomere length are independently associated with chronological age and mortality. Int J Epidemiol (Epub ahead of print, April 13, 2016)Google Scholar
45 : Life stress, glucocorticoid signaling, and the aging epigenome: implications for aging-related diseases. Neurosci Biobehav Rev 2017; 74(Pt B):356–365Crossref, Medline, Google Scholar
46 : Childhood maltreatment predicts unfavorable course of illness and treatment outcome in depression: a meta-analysis. Am J Psychiatry 2012; 169:141–151Link, Google Scholar
47 : Childhood maltreatment and psychopathology: a case for ecophenotypic variants as clinically and neurobiologically distinct subtypes. Am J Psychiatry 2013; 170:1114–1133Link, Google Scholar
48 : Role of epigenetics in the brain, in Epigenetics in Psychiatry. Edited by Avramopoulos D, Grayson DR, Peedicayil J. San Diego, Elsevier, 2014, pp 79–99Crossref, Google Scholar
49 : Adult hippocampal neurogenesis: regulation, functional implications, and contribution to disease pathology. Neurosci Biobehav Rev 2009; 33:232–52Crossref, Medline, Google Scholar
50 : Is depression associated with increased oxidative stress? A systematic review and meta-analysis. Psychoneuroendocrinology 2015; 51:164–175Crossref, Medline, Google Scholar
51 : Major depressive disorder and hypothalamic-pituitary-adrenal axis activity: results from a large cohort study. Arch Gen Psychiatry 2009; 66:617–626Crossref, Medline, Google Scholar
52 : Serum levels of brain-derived neurotrophic factor in major depressive disorder: state-trait issues, clinical features, and pharmacological treatment. Mol Psychiatry 2011; 16:1088–1095Crossref, Medline, Google Scholar
53 : The trajectory of the blood DNA methylome ageing rate is largely set before adulthood: evidence from two longitudinal studies. Age (Dordr) 2016; 38:65Crossref, Medline, Google Scholar



