Association of Protein Phosphatase PPM1G With Alcohol Use Disorder and Brain Activity During Behavioral Control in a Genome-Wide Methylation Analysis
Abstract
Objective:
The genetic component of alcohol use disorder is substantial, but monozygotic twin discordance indicates a role for nonheritable differences that could be mediated by epigenetics. Despite growing evidence associating epigenetics and psychiatric disorders, it is unclear how epigenetics, particularly DNA methylation, relate to brain function and behavior, including drinking behavior.
Method:
The authors carried out a genome-wide analysis of DNA methylation of 18 monozygotic twin pairs discordant for alcohol use disorder and validated differentially methylated regions. After validation, the authors characterized these differentially methylated regions using personality trait assessment and functional MRI in a sample of 499 adolescents.
Results:
Hypermethylation in the 3′-protein-phosphatase-1G (PPM1G) gene locus was associated with alcohol use disorder. The authors found association of PPM1G hypermethylation with early escalation of alcohol use and increased impulsiveness. They also observed association of PPM1G hypermethylation with increased blood-oxygen-level-dependent response in the right subthalamic nucleus during an impulsiveness task.
Conclusions:
Overall, the authors provide first evidence for an epigenetic marker associated with alcohol consumption and its underlying neurobehavioral phenotype.
Alcohol dependence and alcohol abuse are chronically relapsing disorders with a significant public health burden (1). Early escalation of alcohol use among adolescents is a risk factor for future alcohol use disorders (2) and is associated with externalizing disorders (3), which are characterized by high impulsiveness (4). High impulsiveness is itself a well-established risk factor for alcohol use disorders in adolescence (5, 6).
Twin studies indicate that the heritability of alcohol use disorder is 50%–60%, whereas environmental and stochastic effects account for 40%–50% of its variability (7). Investigation of discordant monozygotic twin pairs allows control for genetic variation, enabling an investigation of nonheritable effects (8). It has been suggested that nonheritable influences can be mediated through epigenetic mechanisms (9). Epigenetic processes are essential for normal cellular development and differentiation, and they allow the regulation of gene function through nonmutagenic mechanisms. DNA methylation, a key epigenetic process, is involved in various neurological and cognitive processes, such as neurogenesis (10), brain development (11), learning/memory (12), neurodegeneration (13), and several neuropsychiatric disorders (14). Differential methylation has been observed in peripheral blood and in brain tissue from alcohol-dependent patients, which suggests that epigenetic profiles may be useful as markers for alcohol use disorders (15, 16).
Genome-wide methylation studies show that DNA methylation variability between monozygotic twins accounts for phenotypic discordances (17) in neuropsychiatric disorders, including schizophrenia (18), bipolar disorder (19), and autism spectrum disorder (20). No such investigation has been conducted in alcohol use disorder. While studies have reported associations between DNA methylation patterns and neuropsychiatric disorders, to our knowledge their relation to brain processes has not been investigated.
In this study, we assessed genome-wide DNA methylation in 18 monozygotic twin pairs from the population-based FinnTwin16 study (21) who were discordant for alcohol use disorder, using peripheral blood DNA to identify differentially methylated regions. We investigated behavioral and neuronal processes associated with the most significant differentially methylated region by assessing its association with alcohol-related behavior, personality traits, and brain activation in a functional neuroimaging epigenetics data set of 499 adolescents from the IMAGEN study (www.imagen-europe.com) (22).
Method
FinnTwin16 Study Participants
Twin pairs were part of the FinnTwin16 study, a population-based Finnish cohort of monozygotic and dizygotic twins. They were studied at age 16, with subsequent surveys at ages 17, 18.5, and 24). Among 104 monozygotic twin pairs, 36 were discordant for alcohol dependence or alcohol abuse. From this group, we randomly selected 18 pairs for genome-wide methylation analysis (Table 1; see also Table S1 in the data supplement that accompanies the online edition of this article). Discordant twins were selected using the Rutgers Alcohol Problem Index (23), administered at ages 18.5 and 24, with differences in DSM-III-R symptoms used as additional criteria (assessed using the Semi-Structured Assessment for the Genetics of Alcoholism). We focused on the Rutgers Alcohol Problem Index because in this age group, with a recent and short history of problems, DSM criteria are not sufficiently informative. Scores at ages 18 and 24 were significantly different between individuals with and without alcohol use disorders among the 18 twin pairs (t=−2.552, df=16, p=0.011). At interview at age 24, blood was sampled from both twins, and DNA was extracted using the Gentra Autopure LS System (Gentra Systems, Minneapolis). Monozygosity of twin pairs was confirmed by the Paternity Testing Unit at the Finnish National Public Health Institute. The study protocol was approved by local ethics committees at Helsinki and Indiana Universities.
| Sample and Characteristica | ||
|---|---|---|
| Twin pairs discordant for alcohol use disorder at age 24 | N | % |
| Male | 7 | 38.9 |
| Alcohol dependence diagnosis | 16 | 88.9 |
| Alcohol abuse diagnosis | 2 | 11.1 |
| Mean | SD | |
| Number of DSM-III-R symptoms in the twin without a substance use diagnosis | 1.6 | 0.26 |
| Number of DSM-III-R symptoms in the twin with a substance use diagnosis | 3.5 | 0.22 |
| Adolescent sample | N | % |
| Male | 222 | 44.5 |
| Mean | SD | |
| Development score at age 14 (Pubertal Development Scale) (N=399) | 2.88 | 0.56 |
| Increase in daily amount of drinking (European School Survey Project on Alcohol and Other Drugs) (N=353) | 0.83 | 1.06 |
| Impulsivity at age 16 (Substance Use Risk Profile Scale) (N=399) | 14.02 | 2.75 |
| Sensation seeking at age 16 (Substance Use Risk Profile Scale) (N=399) | 11.48 | 2.02 |
TABLE 1. Characteristics of a Sample of 18 Monozygotic Twin Pairs Discordant for Alcohol Use Disorder and a Sample of 499 Adolescents in a Genome-Wide Methylation Analysisa
Genome-Wide Methylation Assay
Enrichment of unmethylated fraction of genomic DNA (gDNA) was performed by methylation-sensitive restriction of gDNA, amplification, and labeling prior to array hybridization, as previously described (24). (For more details on this and other aspects of the study’s methodology, see the data supplement). Cy3-labeled DNA from each twin was hybridized with Cy5-labeled control DNA to a NimbleGen DNA Methylation 385k Array (Roche NimbleGen, Madison, Wisc.). The array consists of 385,000 60-mer oligonucleotide probes covering promoter regions and cytosine-guanine dinucleotide (CpG) islands. Microarray data were normalized using GenePix Pro 6.0 (Molecular Devices, Sunnyvale, Calif.).
Analysis of the Array
Analysis of raw intensity data was performed using Bioconductor, an open-source software suite in the R statistical software environment (25). Raw intensity data were normalized using quantile normalization implemented in the Ringo package (26). Normalized intensities were log2 transformed, and the ratio between each twin pair was used for analysis. A one-class RankProd analysis was used to identify differentially methylated probes between discordant twins (27). RankProd is based on nonparametric statistics derived from the rank product of each gene. As this procedure is similar to false-discovery-rate estimation, we used percentage of false positives <0.05 for selecting differentially methylated probes.
Verification of Alcohol Use Disorder-Associated Differentially Methylated Regions
Top-ranked alcohol use disorder-associated differentially methylated regions were verified in 18 discordant twin pairs (Table 1) using bisulfite polymerase chain reaction (PCR) and the Sequenom EpiTYPER system (Sequenom, Hamburg). Methylation sites contained in the following genes were assessed: PPMG1, INS-IGF2, FMN1, SEPHS2, SLC6A3, AIM1, OPRL1, and PIPOX. Since we enriched samples for unmethylated fraction of the gDNA using methylation-sensitive restriction enzymes, assays were designed to cover areas containing HpaII and Hin6I cut-sites, adjacent to the 60-mer probes in the NimbleGen DNA Methylation. DNA was bisulfite treated using the EZ-96 DNA methylation kit (Zymo Research, Irvine, Calif.). Primer sequences, PCR conditions, amplicon length, and number of CpG sites are listed in Table S2 in the online data supplement. To detect differentially methylated regions, we performed paired t tests for each single CpG site detectable within each amplicon.
IMAGEN Study Participants
The methylation profile of the 3′-protein-phosphatase-1G (PPM1G) gene locus was investigated in 499 14-year-olds (Table 1) from IMAGEN, a European multicenter imaging-genetic/epigenetic study. Participants were tested in eight assessment centers in four European countries. The Psytools software package (Delosis, London) was used for behavioral characterization. The assessment battery of questionnaires and cognitive tasks was self-administered both online in participants’ homes and at the neuroimaging facilities. The study was approved by local ethics research committees at each site. Written informed consent was obtained from all participants and from their legal guardians. A detailed description of recruitment and assessment procedures as well as inclusion and exclusion criteria has been published previously (22).
Behavioral Characterization
Escalation of daily drinking between ages 14 and 16 was measured by calculating the difference in daily drinking between the two ages using the European School Survey Project on Alcohol and Other Drugs. Externalizing behavioral risk factors for substance abuse or dependence were measured with the Substance Use Risk Profile Scale (28), which assesses independent risks factors, including two externalizing factors, impulsivity and sensation seeking (both the sum of five items). Pubertal development was determined using the Pubertal Development Scale (29), and a development score was obtained by calculating the mean of the scores on the five indicators. Data quality was controlled by context checks administered at the start of each task.
PPM1G Methylation Analysis
DNA was extracted from whole blood samples (∼10 mL) collected at age 14 using the Gentra Puregene Blood Kit (QIAGEN, Valencia, Calif.). DNA was bisulfite treated using the EZ-96 DNA methylation kit, and DNA methylation of PPM1G was assessed using bisulfite PCR and the Sequenom EpiTYPER system.
Stop Signal Task
Data on an event-related stop signal task from 393 14-year-old IMAGEN participants for whom complete data were available were analyzed using the contrast “stop success versus go success” to study neural responses to successful and unsuccessful inhibitory control (30).
Functional MRI Data Acquisition and Analysis
Structural and functional MRI (fMRI) data were acquired with 3-T scanners at eight sites using compatible scanning variables and the same scanning protocol at all sites. Full details of MRI acquisition protocols and quality checks have been described previously, including standardization across MRI scanners (22). Effect of MRI site was controlled for as a nuisance covariate in all statistical analyses. Effect of MRI site and scanner on the magnitude of the fMRI response has been formally evaluated previously (31). All acquired images were preprocessed and analyzed using SPM8 (http://www.fil.ion.ucl.ac.uk/spm) at NeuroSpin (Paris). We extracted regions of interest using the MarsBaR toolbox (http://marsbar.sourceforge.net). The right inferior frontal gyrus was defined according to a human automated anatomical labeling atlas (32), and the right subthalamic nucleus was defined by the Lucerna et al. atlas (33). All regions of interest were extracted to match the stop success versus go success contrast t maps. The center of the resulting right inferior frontal gyrus region of interest was at Montreal Neurological Institute coordinates x=49, y=22, z=16, with a volume of 25,504 mm3. The center of the right subthalamic nucleus region of interest was located at coordinates x=10, y=−15, z=−5, with a volume of 1,000 mm3. Averaged beta values based on all voxels across time series in the regions of interest were used for all analyses.
PPM1G Gene Expression and Genotype Analysis
Total RNA was extracted from whole blood cells collected at age 14 using the PAXgene Blood RNA Kit (QIAGEN). Gene expression profiling was performed by hybridizing labeled cRNA on Illumina HumanHT-12 v4 Expression BeadChips (Illumina, San Diego). Normalized expression data of the probe mapping to PPM1G were extracted, log-transformed, screened for outliers, and tested for association with PPM1G methylation data.
DNA was purified from whole blood samples (∼10 mL) preserved in EDTA tubes using the Gentra Puregene Blood Kit. Genotype information was collected at 582,982 markers using the Illumina HumanHap610 Genotyping BeadChip. Genotype data from four single-nucleotide polymorphisms (SNPs), present within 50 kb of the PPM1G CpG site analyzed (rs7602534, rs11675428, rs704791, and rs1260342), were tested for association with PPM1G methylation. Data for rs2384629, previously associated with alcohol use disorders (34), were imputed, and haplotypes covering the gene were calculated using Plink (http://pngu.mgh.harvard.edu/∼purcell/plink/).
Association Analyses
The general linear model was used to determine associations among methylation of a CpG site present in the 3′ untranslated region (3′-UTR) of PPM1G, found differentially methylated in the twins discordant for alcohol use disorders, as well as PPM1G gene expression and genotype, quantity and frequency of drinking, escalation of daily drinking, impulsiveness, and stop signal task blood-oxygen-level-dependent (BOLD) responses. Gender, puberty, and study site were controlled for in all analyses. Handedness was controlled for in association analyses of BOLD activation. Corrected p values (pcorrected) were computed using 10,000 permutations. Partial eta-squared (η2p), a measure of effect size calculated after removing the contribution from other control variables, is reported for all associations.
Results
Genome-Wide Analysis of Differentially Methylated Genes in Monozygotic Twins Discordant for Alcohol Use Disorder
Genome-wide DNA methylation was first assessed by microarray analysis in 18 monozygotic twin pairs discordant for alcohol use disorder (Table 1) of 385,000 60-mer oligonucleotide probes covering both promoter regions and CpG islands. Using a nonparametric method and a cutoff of the estimated percentage of false positive predictions <0.05 and a p value <1×10−5, we identified 77 differentially methylated regions that exhibit an alcohol use disorder-associated differential methylation (see Table S3 in the online data supplement).
The majority of the 77 differentially methylated regions (68%) were hypermethylated in the affected twin, consistent with previous findings showing an association between DNA hypermethylation and alcohol use disorder (15). The identified regions were associated with 62 genes, with 57% located in the gene body, 30% in promoters, and the rest intergenic (see Figure S1A in the data supplement). The majority of differentially methylated regions (65%) were located outside of CpG-rich regions (CpG islands), with 41% of the sites located in shores flanking the islands, which have been implicated in gene regulation (35) (see Figure S1B in the data supplement).
The most significant differentially methylated regions located within genes (body and promoter) were divided into hypermethylated and hypomethylated regions; the top 10 differentially methylated regions of each category are listed in Table 2. Among these regions, we selected those with an absolute delta (Δ) DNA methylation intensity value (the mean difference in DNA methylation intensity between the affected and the unaffected member in each twin pair) higher than 0.5 for technical replication using mass spectrometry. These included differentially methylated regions in the PPMG1, INS-IGF2, FMN1, SEPHS2, SLC6A3, AIM1, OPRL1, and PIPOX genes, which we analyzed in the 18 monozygotic twin pairs with a Sequenom EpiTYPER (see Table S2 in the data supplement). In the twins with an alcohol use disorder, we found significant increases in percent methylation of the CpG site in the 3′-UTR of PPM1G (p=0.005, mean Δ=2.4%) and in a CpG site contained in the OPRL1 probe (p=0.017, mean Δ=3.6%), confirming the findings from the microarray. We were unable to confirm differential DNA methylation in the other regions, possibly because the Sequenom assay could not interrogate the specific methylation-sensitive restriction enzyme cut-site driving the difference reported in the microarray.
| Gene (Nimblegen Identification Number) | Gene Symbol | pfpa | Location | CpGb Context |
|---|---|---|---|---|
| Top hypermethylated regions (p<1×10–5) | ||||
| Protein phosphatase 1G (CHR02P027457864) | PPM1G | <1×10–5 | Body | Shore |
| Insulin- insulin-like growth factor 2 (CHR11P002137905) | INS_IGF2 | <1×10–5 | Body | Shelf |
| NA (CHR05P064022468) | NA | 3×10–4 | Intergenic | Island |
| Nucleosome assembly protein 1-like 2 (CHRXP072351512) | NAP1L2 | 3×10–3 | Body | Open sea |
| Solute carrier family 6 (neurotransmitter transporter, dopamine), member 3 (CHR05P001469753) | SLC6A3 | 4×10–3 | Body | Shore |
| Hypothetical protein LOC149840 (CHR20P005679048) | NA | 4×10–3 | Body | Open sea |
| Ubiquitin-conjugating enzyme E2M pseudogene 1 (CHR16P034261914) | UBE2MP1 | 6×10–3 | Body | Shore |
| Absent in melanoma 1 (CHR06P107065946) | AIM1 | 7×10–3 | Body | Shore |
| NA (CHR09P069075112) | NA | 7×10–3 | Intergenic | Island |
| Opioid receptor-like 1 (CHR20P062188944) | OPRL1 | 7×10–3 | Body | Island |
| Top hypomethylated regions (p<1×10–5) | ||||
| Neurotrophic tyrosine kinase, receptor, type 1 (CHR01P155095309) | NTRK1 | >1×10–5 | Body | Shore |
| NA (CHR02P095678989) | NA | >1×10–5 | Intergenic | Shore |
| Formin 1 (CHR15P031274371) | FMN1 | >1×10–5 | Body | Island |
| Selenophosphate synthetase 2 (CHR16P030364503) | SEPHS2 | >1×10–5 | Body | Island |
| Myocyte enhancer factor 2D (CHR01P154736441) | MEF2D | 3×10–4 | Body | Island |
| 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (CHR10P006245627) | PFKFB3 | 3×10–4 | Body | Island |
| Solute carrier family 45, member 4 (CHR08P142298123) | SLC45A4 | 4×10–4 | Body | Island |
| L-pipecolic acid oxidase (CHR17P024393350) | PIPOX | 4×10–4 | Body | Open sea |
| Fibroblast growth factor (acidic) intracellular binding protein (CHR11P065412364) | FIBP | 2×10–3 | Body | Island |
| MIRLET7B host gene (nonprotein coding) (CHR22P044859950) | MIRLET7BHG | 2×10–3 | Body | Open sea |
TABLE 2. Differentially Methylated Regions in the Twin With an Alcohol Use Disorder Compared With the Unaffected Twin
Association of PPM1G Methylation, Gene Expression, and Genotype
As the PPM1G differentially methylated region showed the most significant association with alcohol use disorders, both in genome-wide analysis and in technical replication, we proceeded with a more detailed analysis and validation of PPM1G in an independent sample of 499 adolescents from the IMAGEN imaging-genetics study (Table 1) for whom gene expression and DNA methylation data were available. A linear regression analysis of PPM1G methylation and PPM1G gene expression revealed significant association (t=−1.93, p=0.027 [one-tailed]; η2p=0.007), with higher methylation levels associated with lower mRNA levels (see Figure S2 in the data supplement).
To rule out possible genotype effects on methylation, we performed a linear regression of PPM1G methylation and five SNPs, including rs2384629, which was previously reported to be associated with alcohol dependence (37). The other SNPs covering the PPM1G locus were rs7602534, rs11675428, rs704791, and rs1260342. Neither a single SNP nor a haplotype analysis (±50 kb of the PPM1G CpG site analyzed) yielded significant association with methylation of PPM1G (see Table S4 in the data supplement).
Association of PPM1G Methylation With Drinking Escalation and Trait Impulsiveness
To investigate whether the differentially methylated CpG site in PPM1G was associated with degree of alcohol exposure, we explored in the IMAGEN sample the association of PPM1G methylation and drinking measures at age 14. We found no association of PPM1G methylation and quantity (t=−0.45, p=0.65; η2p =0.0004) or frequency of drinking during the 12 months prior to blood acquisition (t=−0.15, p=0.88; η2p =0.00005) or during the month prior to blood acquisition (t=–0.86, p=0.39; η2p=0.0017) (see Table S5 in the data supplement). This suggests that PPM1G hypermethylation is not caused by alcohol exposure at age 14. As escalation of alcohol use in adolescents is associated with increased risk for alcohol-related problems and alcohol use disorders (28), we explored a possible association of PPM1G methylation with escalation in daily use of alcohol between ages 14 and 16. We selected among the 499 adolescents of the IMAGEN study those who reported drinking at ages 14 or 16 (N=352) and found that PPM1G hypermethylation was positively associated with escalation of daily drinking (t=2.17, p=0.015 [one-tailed], pcorrected=0.024; η2p =0.014) (Figure 1A). Next, we investigated a possible association of escalation of alcohol use with externalizing behavioral risk factors for alcohol use disorders (36). Escalation of daily drinking was associated with impulsiveness (t=2.33, p=0.020, pcorrected=0.04; η2p=0.017) (Figure 1B) but not with sensation seeking. Impulsiveness, in turn, was associated with PPM1G methylation (t=2.56, p=0.012, pcorrected=0.032; η2p =0.016; N=499) (Figure 1C), indicating a role of PPM1G methylation in both risky drinking and behavioral risk factors for alcohol use disorders in adolescents. When we controlled for impulsiveness, we lost the significance of the association of PPM1G methylation and escalation of daily drinking (p=0.065) and observed a decrease in effect size by 50% (with the η2p value decreasing from 0.014 to 0.007). There was no association of the previously reported PPM1G SNP rs2384629 (34) with escalation of drinking or with trait impulsiveness (see Table S4 in the data supplement).
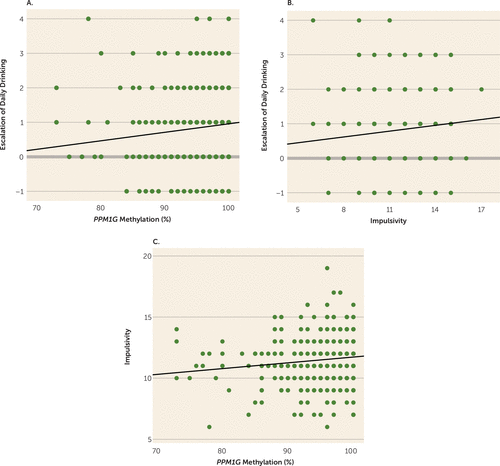
FIGURE 1. Association of PPM1G Methylation With Drinking Escalation (N=352) and Trait Impulsiveness (N=399) in Adolescentsa
a As shown in panel A, PPM1G methylation is positively associated with an increase in amount of daily drinking (corrected p=0.024). PPM1G methylation at age 14 predicts an increase in daily drinking between ages 14 and 16. In panel B, an increase in amount of daily drinking is positively associated with impulsiveness in 16-year-old adolescents (corrected p=0.041). In panel C, PPM1G methylation is positively associated with impulsiveness (corrected p=0.032). All graphs represent data for each individual and have linear fit lines. In panels A and B, escalation of daily drinking is measured as the difference between age 14 and age 16 on an item on amount of daily drinking from the European School Survey Project on Alcohol and Other Drugs questionnaire (on a five-level ordinal scale). In panels B and C, impulsiveness is represented by a sum score of five items (on a five-level ordinal scale) of the Substance Use Risk Profile Scale.
Association of PPM1G Methylation and Activation of the Subthalamic Nucleus During a Behavioral Inhibition Task
We examined a possible association between PPM1G methylation and brain activity during functional neuroimaging registration of the stop signal task, which assesses behavioral (motor) inhibition and is associated with alcohol use disorders (37, 38). In humans, voluntary inhibition of manual movements relies on a right-lateralized frontal–basal ganglia–thalamic pathway (39). This network includes the inferior frontal gyrus, which revokes planned movements through the subthalamic nucleus (40, 41) of the right hemisphere. We selected these regions of interest in the stop success versus go success contrast. In 14-year-olds (N=393) we found a positive association between PPM1G methylation and right subthalamic nucleus BOLD response (t=2.25, p=0.021, pcorrected=0.038; η2p=0.013) (Figure 2A,B) but not with right inferior frontal gyrus BOLD response. There was no association of the previously reported PPM1G SNP rs2384629 (34) with activation of the right subthalamic nucleus (see Table S4 in the data supplement).
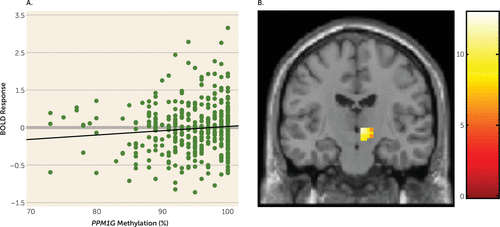
FIGURE 2. Association of PPM1G Methylation and Activation of the Subthalamic Nucleus During a Stop Signal Task in Adolescents (N=393)a
a As shown in panel A, PPM1G methylation is positively associated with blood-oxygen-level-dependent (BOLD) activation in the right subthalamic nucleus in 14-year-olds (corrected p=0.038). The graph depicts the percentage of PPM1G methylation and averaged beta values of activation of the right subthalamic nucleus for each individual; a linear fit line was added. In panel B, a coronal section shows methylation differences in activation of the right subthalamic nucleus (t value of activation, with a lighter color indicating a stronger activation) during successful inhibition, indicating an association between PPM1G methylation and activation of the right subthalamic nucleus (coordinates: x=10, y=−15, z=−5).
Discussion
In a genome-wide study of DNA methylation in adult monozygotic twin pairs discordant for alcohol use disorders, we discovered association of hypermethylation of a differentially methylated region in the 3′-UTR of PPM1G with alcohol use disorder. PPM1G is a member of the PP2C family of serine/threonine protein phosphatases and has previously been linked with alcohol dependence (34). In an independent population-based sample of the IMAGEN cohort, we showed association of increased DNA methylation in PPM1G with early escalation of drinking and impulsiveness, both risk factors for future alcohol use disorders (5, 6). We did not find association with quantity or frequency measures in the IMAGEN sample at age 14; neither did we see an association of this differentially methylated region with amount of alcohol consumption in twins at the time of blood acquisition at age 25 (data not shown). Together with low levels of alcohol consumption at the time of the neuroimaging registration at age 14, these observations suggest that PPM1G methylation influences impulsiveness, which in turn may result in problematic patterns of drinking and behavior, as opposed to being a marker for the amount of alcohol consumption. This distinction is supported by previous data showing that the genetics of alcohol use disorders are distinct from the genetics of consumption, implicating different underlying biological pathways (42). We did not find association of PPM1G genotype and methylation profile, suggesting that previously reported genetic associations in PPM1G (34) and the differential methylation of PPM1G are independent.
We speculate that differential methylation of PPM1G may be a result of unshared environmental factors, which we were unable to identify. Several nonfamilial risk factors have been associated with alcohol use disorders, especially during adolescence, including peer influence and affiliation with drinking peers (43, 44). It would be intriguing to characterize environmental factors more comprehensively to identify those associated with PPM1G methylation.
PPM1G methylation is associated with greater activation of the right subthalamic nucleus in the stop signal task when measuring stop success. This points toward an increased effort to carry out successful behavioral inhibition in those individuals with increased methylation of PPM1G. The subthalamic nucleus is a basal ganglia structure whose functions include transmission of inhibitory cortical signals to basal ganglia, including the striatum (46, 50, 51). Several studies have suggested that the right subthalamic nucleus and right inferior frontal gyrus are correlated with response inhibition, suggesting that they form a frontal-subcortical pathway of control inhibition (40, 45, 46). The right-lateralized subthalamic nucleus and inferior frontal gyrus have been used as regions of interest for response inhibition tasks (i.e., the stop signal task and go/no-go task) in studies reporting compromised impulse control in alcohol-dependent individuals (37, 38) and reduced ability to cancel prepotent responses (47), as well as in lesion studies (48), deep brain stimulation studies (49), and fMRI studies (50) of response inhibition in individuals with Parkinson’s disease. Studies of in vivo deep brain stimulation of the subthalamic nucleus in Parkinson’s patients (51) and in animal models (52) have shown a decrease in craving and regulation of substance preference, indicating the involvement of this brain region in neural processes that underlie addictive behavior.
While PPM1G methylation is independently associated with both trait impulsiveness (measured by the Substance Use Risk Profile Scale) and right subthalamic nucleus activation in the stop signal task, we found no association between trait impulsiveness and right subthalamic nucleus activation (data not shown). This is unsurprising, as impulsive traits do not necessarily correlate with behavioral indices of impulsiveness (53), which have been shown to load into independent principal components (54).
PPM1G is expressed in different brain areas, including in the subthalamic nucleus, where its gene product is thought to be involved in the dephosphorylation of metabotropic glutamate receptors (mGluR2/3) (57, 58), thus potentially regulating the transmission of cortical signals to the striatum (46, 50, 51).
Increased methylation of PPM1G is associated with reduced mRNA levels. Although these data were ascertained from whole blood and do not necessarily reflect gene regulation in specific brain areas at a particular age, they indicate that PPM1G methylation has the capacity to influence gene expression levels. Its localization in the 3′-UTR suggests that differential methylation could influence mRNA termination and stability (55). This is in line with evidence demonstrating the importance of intragenic methylation outside of canonical CpG islands and promoter regions in regulating transcriptional activity. For example, intragenic methylation can act as an alternative promoter or it can regulate splicing (55). The PPM1G differentially methylated region is located in a shore flanking a CpG island. Similar localizations were observed in most differentially methylated regions identified in the genome-wide methylation study. Analogous to observations in cancer studies (35), this indicates that methylation in shores could be important in gene regulation and the pathophysiology of psychiatric disorders.
As methylation patterns and their regulation diverge not only across tissues and cell populations (56) but also between brain regions (57) and across the lifespan (58), a definitive validation study would have to be conducted in the subthalamic nucleus of adolescent postmortem brains. There are, however, no such samples in sufficient quantities to support an association analysis of methylation and gene expression.
In the monozygotic discordant twin pairs, there are no data available beyond age 24. Since both methylation patterns (58) and alcohol use disorder diagnosis (59) can change over the lifespan, it would be interesting to study discordant twin pairs at a later age to follow the trajectories of the disorder and their association with PPM1G methylation.
In conclusion, we identified a differentially methylated region in the PPM1G gene associated with alcohol use disorders in adults, and we provide evidence for its association with brain mechanisms and behaviors that underlie risk in adolescents for future alcohol-related problems. Our data indicate that methylation of PPM1G may influence behavioral inhibition by altering activity of the right subthalamic nucleus, which integrates neural signals necessary for behavioral control.
1 : Global burden of disease attributable to mental and substance use disorders: findings from the Global Burden of Disease Study 2010. Lancet 2013; 382:1575–1586Crossref, Medline, Google Scholar
2 : A developmental perspective on alcohol and youths 16 to 20 years of age. Pediatrics 2008; 121(suppl 4):S290–S310Crossref, Medline, Google Scholar
3 : Genetic and environmental predictors of latent trajectories of alcohol use from adolescence to adulthood: a male twin study. Alcohol Clin Exp Res 2013; 37:498–506Crossref, Medline, Google Scholar
4 : Neurocognitive endophenotypes of impulsivity and compulsivity: towards dimensional psychiatry. Trends Cogn Sci 2012; 16:81–91Crossref, Medline, Google Scholar
5 : Determinants of early alcohol use in healthy adolescents: the differential contribution of neuroimaging and psychological factors. Neuropsychopharmacology 2012; 37:986–995Crossref, Medline, Google Scholar
6 : Adolescent impulsivity phenotypes characterized by distinct brain networks. Nat Neurosci 2012; 15:920–925Crossref, Medline, Google Scholar
7 : Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med 1997; 27:1381–1396Crossref, Medline, Google Scholar
8 : The continuing value of twin studies in the omics era. Nat Rev Genet 2012; 13:640–653Crossref, Medline, Google Scholar
9 : Drugs and addiction: an introduction to epigenetics. Addiction 2011; 106:480–489Crossref, Medline, Google Scholar
10 : Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat Neurosci 2010; 13:1338–1344Crossref, Medline, Google Scholar
11 : Global epigenomic reconfiguration during mammalian brain development. Science 2013; 341:1237905Crossref, Medline, Google Scholar
12 : TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron 2013; 79:1086–1093Crossref, Medline, Google Scholar
13 : Epigenetic mechanisms in neurological disease. Nat Med 2012; 18:1194–1204Crossref, Medline, Google Scholar
14 : From promises to practical strategies in epigenetic epidemiology. Nat Rev Genet 2013; 14:585–594Crossref, Medline, Google Scholar
15 : Genome-wide DNA methylation analysis in alcohol dependence. Addict Biol 2013; 18:392–403Crossref, Medline, Google Scholar
16 : Global DNA promoter methylation in frontal cortex of alcoholics and controls. Gene 2012; 498:5–12Crossref, Medline, Google Scholar
17 : DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet 2009; 41:240–245Crossref, Medline, Google Scholar
18 : Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet 2011; 20:4786–4796Crossref, Medline, Google Scholar
19 : Aberrant DNA methylation associated with bipolar disorder identified from discordant monozygotic twins. Mol Psychiatry 2008; 13:429–441Crossref, Medline, Google Scholar
20 : Methylomic analysis of monozygotic twins discordant for autism spectrum disorder and related behavioural traits. Mol Psychiatry 2014; 19:495–503Crossref, Medline, Google Scholar
21 : Twin studies in Finland 2006. Twin Res Hum Genet 2006; 9:772–777Crossref, Medline, Google Scholar
22 : The IMAGEN study: reinforcement-related behaviour in normal brain function and psychopathology. Mol Psychiatry 2010; 15:1128–1139Crossref, Medline, Google Scholar
23 : Towards the assessment of adolescent problem drinking. J Stud Alcohol 1989; 50:30–37Crossref, Medline, Google Scholar
24 : Microarray-based DNA methylation profiling: technology and applications. Nucleic Acids Res 2006; 34:528–542Crossref, Medline, Google Scholar
25 : Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5:R80Crossref, Medline, Google Scholar
26 : Ringo: an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics 2007; 8:221Crossref, Medline, Google Scholar
27 : Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 2004; 573:83–92Crossref, Medline, Google Scholar
28 : The Substance Use Risk Profile Scale: a scale measuring traits linked to reinforcement-specific substance use profiles. Addict Behav 2009; 34:1042–1055Crossref, Medline, Google Scholar
29 : Adolescent development. Annu Rev Psychol 1988; 39:583–607Crossref, Medline, Google Scholar
30 : Mapping motor inhibition: conjunctive brain activations across different versions of go/no-go and stop tasks. Neuroimage 2001; 13:250–261Crossref, Medline, Google Scholar
31 : Creating probabilistic maps of the face network in the adolescent brain: a multicentre functional MRI study. Hum Brain Mapp 2012; 33:938–957Crossref, Medline, Google Scholar
32 : Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002; 15:273–289Crossref, Medline, Google Scholar
33 : In Vivo Atlas of Deep Brain Structures. Berlin, Springer-Verlag, 2002Crossref, Google Scholar
34 : Evidence for genes on chromosome 2 contributing to alcohol dependence with conduct disorder and suicide attempts. Am J Med Genet B Neuropsychiatr Genet 2010; 153B:1179–1188Medline, Google Scholar
35 : Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells, and fibroblasts. Nat Genet 2009; 41:1350–1353Crossref, Medline, Google Scholar
36 : Development of the Alcohol Use Disorders Identification Test (AUDIT): WHO Collaborative Project on Early Detection of Persons With Harmful Alcohol Consumption, II. Addiction 1993; 88:791–804Crossref, Medline, Google Scholar
37 : Altered impulse control in alcohol dependence: neural measures of stop signal performance. Alcohol Clin Exp Res 2009; 33:740–750Crossref, Medline, Google Scholar
38 : The role of behavioral impulsivity in the development of alcohol dependence: a 4-year follow-up study. Alcohol Clin Exp Res 2008; 32:1681–1687Crossref, Medline, Google Scholar
39 : Converging evidence for a fronto-basal-ganglia network for inhibitory control of action and cognition. J Neurosci 2007; 27:11860–11864Crossref, Medline, Google Scholar
40 : Cortical and subcortical contributions to stop signal response inhibition: role of the subthalamic nucleus. J Neurosci 2006; 26:2424–2433Crossref, Medline, Google Scholar
41 : Stimulation of the subthalamic region facilitates the selection and inhibition of motor responses in Parkinson’s disease. J Cogn Neurosci 2006; 18:626–636Crossref, Medline, Google Scholar
42 : Measures of current alcohol consumption and problems: two independent twin studies suggest a complex genetic architecture. Alcohol Clin Exp Res 2011; 35:2152–2161Crossref, Medline, Google Scholar
43 : Childhood and adolescent predictors of heavy episodic drinking and alcohol use disorder at ages 21 and 33: a domain-specific cumulative risk model. J Stud Alcohol Drugs 2014; 75:684–694Crossref, Medline, Google Scholar
44 : Parent, sibling, and peer associations with subtypes of psychiatric and substance use disorder comorbidity in offspring. Drug Alcohol Depend 2013; 128:20–29Crossref, Medline, Google Scholar
45 : Stop-signal inhibition disrupted by damage to right inferior frontal gyrus in humans. Nat Neurosci 2003; 6:115–116Crossref, Medline, Google Scholar
46 : Subcortical processes of motor response inhibition during a stop signal task. Neuroimage 2008; 41:1352–1363Crossref, Medline, Google Scholar
47 : Disorder-specific dysfunction in right inferior prefrontal cortex during two inhibition tasks in boys with attention-deficit hyperactivity disorder compared to boys with obsessive-compulsive disorder. Hum Brain Mapp 2010; 31:287–299Crossref, Medline, Google Scholar
48 : The subthalamic nucleus and inhibitory control: impact of subthalamotomy in Parkinson’s disease. Brain 2014; 137:1470–1480Crossref, Medline, Google Scholar
49 : Effects of deep brain stimulation of the subthalamic nucleus on inhibitory and executive control over prepotent responses in Parkinson’s disease. Front Syst Neurosci 2013; 7:118Crossref, Medline, Google Scholar
50 : Subthalamic nucleus activity dissociates proactive and reactive inhibition in patients with Parkinson’s disease. Neuroimage 2014; 91:273–281Crossref, Medline, Google Scholar
51 : Deep brain stimulation for addiction: why the subthalamic nucleus should be favored. Curr Opin Neurobiol 2013; 23:713–720Crossref, Medline, Google Scholar
52 : Reducing the desire for cocaine with subthalamic nucleus deep brain stimulation. Proc Natl Acad Sci USA 2010; 107:1196–1200Crossref, Medline, Google Scholar
53 : Dimensions of impulsive behavior: personality and behavioral measures. Pers Individ Dif 2006; 40:305–315Crossref, Google Scholar
54 : The relationship between impulsive choice and impulsive action: a cross-species translational study. PLoS ONE 2012; 7:e36781Crossref, Medline, Google Scholar
55 : Functions of DNA methylation: islands, start sites, gene bodies, and beyond. Nat Rev Genet 2012; 13:484–492Crossref, Medline, Google Scholar
56 : Large-scale methylation analysis of human genomic DNA reveals tissue-specific differences between the methylation profiles of genes and pseudogenes. Hum Mol Genet 2000; 9:2651–2663Crossref, Medline, Google Scholar
57 : Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol 2012; 13:R43Crossref, Medline, Google Scholar
58 : Epigenetic variation during the adult lifespan: cross-sectional and longitudinal data on monozygotic twin pairs. Aging Cell 2012; 11:694–703Crossref, Medline, Google Scholar
59 : Characterizing alcohol dependence: transitions during young and middle adulthood. Exp Clin Psychopharmacol 2006; 14:228–244Crossref, Medline, Google Scholar



