Neurocognitive Endophenotypes in a Multiplex Multigenerational Family Study of Schizophrenia
Abstract
Objective: Genetic factors contribute to the development of schizophrenia where cognitive dysfunction is a hallmark. The purpose of this article was to examine computerized neurocognitive measures as candidate endophenotypic markers of liability for schizophrenia in a genetically informative cohort. Method: European Americans from 35 multiplex multigenerational families (N=349) and healthy participants (N=154) underwent clinical assessments and neurocognitive measurements and provided blood samples. The neurocognitive measures included performance (accuracy and speed) from a computerized battery that assessed abstraction/mental flexibility; attention; verbal, face, and spatial memory; spatial processing; sensorimotor processing; and emotion intensity discrimination. Results: Probands, relatives, and comparison subjects differed from each other in performance. Probands demonstrated greatest impairment relative to comparison subjects, followed by family members. Liability for schizophrenia affected the speed-accuracy tradeoff differently for specific neurocognitive domains. Significant heritability estimates were obtained for accuracy of verbal, facial, and spatial memory and spatial and emotion processing. For speed, estimates of heritability were significant for abstraction/mental flexibility, attention, face memory, and spatial and sensorimotor processing. Conclusions: In a multigenerational multiplex design, the authors demonstrated that neurocognitive measures are associated with schizophrenia, differentiate unaffected relatives from comparison subjects, and may have significant presumed heritability. Therefore, they are endophenotypes suitable for genetic studies. Accuracy and speed can be differentially sensitive to presumed genetic liability.
Schizophrenia is a complex disorder with several replicated genetic linkages reported (1) . Since disordered cognitive functioning is a hallmark of schizophrenia, progress in understanding its pathophysiology mandates integration of genetic and neurobiological research. Consistent with anatomic and physiologic findings of frontotemporal dysfunction, neurocognitive measures have indicated deficits in executive domains, learning, and memory (2 , 3) . The potential of neurocognitive measures as markers of genetic liability is supported by studies showing intermediate deficits in attention and memory in unaffected relatives (4 – 9) .
Given the heterogeneity of schizophrenia at the phenotypic and likely genotypic levels, analyzing neurobiological phenotypes may improve power to detect susceptibility loci by constraining some heterogeneity. The possibility that endophenotypes are genetically simpler than disease endpoints is one of their advantages. Furthermore, these quantitative parameters can be measured in family members, where a clinical diagnosis may be absent or difficult to establish. Another advantage is that continuous quantitative traits have inherently more resolution than dichotomous traits. Most important, however, cognitive traits are increasingly being linked to neural systems that will provide more direct mechanistic windows, eventually permitting subcategorization of schizophrenia based on differences in pathophysiology.
The present study examines quantitative neurocognitive measures as candidate endophenotypic markers in multiplex multigenerational families. Our approach requires a different ascertainment strategy from that used in most syndrome-based phenotyping for genetic analysis (10) . Specifically, the power to detect genes for quantitative traits through linkage analyses increases with family size (11) , making extended multigenerational families rather than sibpairs the cohort unit of choice. Because the endophenotypes can be measured in unaffected family members, smaller cohort sizes of probands are necessary. If neurocognitive deficits are associated with genetic liability, they should increase with presumed genetic loading for schizophrenia. Investigations with simplex and multiplex families have supported an additive model in which increased genetic risk is accompanied by increased impairment in language (12) , intelligence, verbal memory, visual reproduction (13) , visual working memory (14) , verbal learning, delayed visual recall, and perceptual- and pure-motor speed (15) . In this first report, we characterize the neurocognitive profile of multiplex multigenerational families with schizophrenia and provide heritability estimates of neurocognitive measures.
Method
Written informed consent was obtained after the procedures had been fully explained. In the case of children (<18), the child’s assent and parental consent were obtained.
Participants
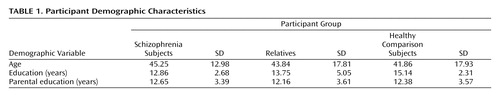
The cohort included 349 European Americans from 35 multiplex multigenerational families (average family size 10.51 [SD=8.46] members, range: 3–32), who met inclusion criteria. A normative group included 154 medically and psychiatrically healthy European Americans ages 19 to 84. Comparison subjects, patients (N=58), and relatives (N=291) did not differ significantly in age and parental education. As expected, patients attained less education than family members and comparison subjects ( Table 1 ). The male:female ratio was as follows: schizophrenia group: 34:24, relatives: 144:147, and comparison subjects: 76:78.
The mode of ascertainment was population based. Potential participants were identified through mental health and consumer organizations in Pennsylvania and bordering states. Suitability for the study was determined based on specified inclusion and exclusion criteria established by standardized screening and assessment. Healthy comparison subjects were recruited from the same communities as probands and families.
Participating probands were older than 18 years and could provide signed informed consent. They met consensus best-estimate DSM-IV diagnosis of schizophrenia and had at least one first-degree affected family member with schizophrenia or schizoaffective disorder, depressed type. In addition, they had an extended multigenerational family, with at least 10 first- and second-degree relatives. Potential probands were excluded if they did not provide written consent to contact family members, their psychosis was linked to substance-related disorders by DSM-IV criteria, they had mental retardation (IQ<70), they had a history of a medical disorder or they were receiving medication that may cause psychosis or neurocognitive deficits, or they were not proficient in English.
Participating family members were older than 15 years and could provide signed informed consent. Family members were excluded if they had mental retardation (IQ<70), a CNS disorder that may render neurocognitive measures noninterpretable, or were not proficient in English. The exclusion criteria applied to the neurocognitive measures, but if diagnosis was established, blood samples were obtained. Potential healthy comparison participants underwent standard screening procedures followed by the same assessment procedure to establish the absence of axis I and cluster A axis II disorders. They were psychiatrically, medically, and neurologically healthy, receiving no psychotropic medications, and reported no first-degree relative with psychosis or mood disorder.
Procedures
Diagnostic assessment
The psychiatric evaluation included the Diagnostic Interview for Genetic Studies, version 2.0 (16) , the Family Interview for Genetic Studies (17) , and review of medical records. The Diagnostic Interview for Genetic Studies was always conducted in person, and the Family Interview for Genetic Studies was conducted in person with at least two family members and, if necessary, by phone when another family member was particularly informative. Interviews were conducted by trained interviewers with established reliability and under the supervision of investigators. A summary statement narrated the history, interview, mental status, examples of answers, and observations.
Two investigators who had not evaluated the individual reviewed each case independently and provided DSM-IV multiaxial lifetime diagnoses. Subjects with psychotic features or disagreement between the investigators were presented in consensus conference, and complex cases were discussed between sites. At each site, interrater reliability among investigators and interviewers was tested at regular intervals using videotaped interviews and bimonthly joint interviews. The interviewers viewed 10 videotaped Diagnostic Interview for Genetic Studies evaluations exchanged between the University of Pennsylvania and the University of Pittsburgh, maintaining kappa values >0.8. The teams met twice a year for diagnostic, reliability, and training purposes.
Neurocognitive measures
Participants were administered a computerized neurocognitive “scan” previously applied to healthy individuals (18) and patients with schizophrenia (19) . It is an efficient test battery administered by research assistants using desktop or portable computers. The battery, designed for large-scale studies, includes a training module and has automated scoring with direct data downloading. The battery assesses the following eight domains:
Abstraction and mental flexibility . The Penn Conditional Exclusion Test (20) presents four objects at a time, and the participant selects the object that does not belong with the other three based on one of three sorting principles. Sorting principles change, and feedback guides their identification (time: 12 minutes).
Attention . The Penn Continuous Performance Test (21) uses a continuous performance test paradigm where the participant responds to seven-segment displays whenever they form a digit. Working memory demands are eliminated because the stimulus is present (time: 8 minutes).
Verbal memory . The Penn Word Memory Test (22) presents 20 target words followed by an immediate recognition trial with targets interspersed with 20 distractors equated for frequency, length, concreteness, and low imageability using Paivio’s norms. Delayed recognition is measured at 20 minutes (time: 4 minutes).
Face memory . The Penn Face Memory Test (22) presents 20 digitized faces subsequently intermixed with 20 foils equated for age, gender, and ethnicity. Participants indicate whether or not they recognize each face immediately and at 20 minutes (time: 4 minutes).
Spatial memory . The Visual Object Learning Test (23) presents 20 Euclidean shapes subsequently interspersed with foils immediately and at 20 minutes (time: 4 minutes).
Spatial processing . Judgment of Line Orientation (24) is a computer adaptation of Benton’s test. Participants see two lines at an angle and indicate the corresponding lines on a simultaneously presented array (time: 6 minutes).
Sensorimotor dexterity . The participant uses a mouse to click on squares appearing at varied locations on the screen (18) . The stimuli become progressively smaller (time: 2 minutes).
Emotion processing . Identification of facial affect was tested with a 40-item Emotion Intensity Discrimination Test (25) . Each stimulus presents two faces of the same individual showing the same emotion (happy or sad) with different intensities. The participant selects the more intense expression. Sets were balanced for gender, age, and ethnicity (5 minutes).
Administration and scoring
The battery was administered in a fixed order using clickable icons. Its administration took about 60 minutes. All except 33 participants yielded valid data for all measures. Missing data occurred because of technical difficulties or failure to follow instructions. Raw scores were converted to z scores using the comparison group mean and then averaged to obtain domain scores. The following two performance indices were calculated: 1) accuracy—the number of correct responses—and 2) speed—the median reaction time for correct responses. Only speed was examined for the sensorimotor dexterity domain because 75% of participants achieved perfect accuracy.
Statistical Analyses
Differences in neurocognition among groups were analyzed by hierarchical linear modeling (26) using the SAS PROC MIXED routine (27) . Family data consist of two hierarchical or multilevel units: participants (level 1) are nested within families (level 2). Because family members are not independent observations, the assumption of independence in analysis of variance (ANOVA)-based methods is violated in the presence of hierarchical data. In practice, we have found endophenotype deficits in family members to be robust to this violation (28 , 29) . However, hierarchical linear modeling formally addresses this problem by modeling the interdependence among members of the same family through testing for a random effect for families (30) . Moreover, hierarchical linear modeling provides more flexibility than ANOVA-based methods in the face of missing data (e.g., scores on a particular task); participants with missing data are not eliminated from analyses (26) .
First, multivariate hierarchical linear modeling analyses were conducted for accuracy and speed to examine overall group effects (schizophrenia, relative, comparison group) and group-by-domain (abstraction and mental flexibility, attention, verbal memory, face memory, spatial memory, spatial processing, sensorimotor dexterity, emotion processing) interactions. Second, for any significant overall effect, pairwise (schizophrenia versus comparison group, relative versus comparison group, schizophrenia versus relative group) multivariate hierarchical linear modeling analyses were conducted to determine pairwise differences. Finally, univariate post hoc hierarchical linear modeling analyses were conducted to determine specific cognitive domains in which groups differed. For these analyses, we also examined family-by-diagnosis interactions on the domain scores.
To maximize power and generalizability, the initial analyses included all ascertained relatives unaffected with schizophrenia, regardless of other axis I or II diagnoses. However, the inclusion of more distant relatives is expected to dilute the appearance of a deficit because of the increased genetic distance from a person with schizophrenia. Consequently, we also compared first-degree relatives with more distant relatives. To address the potential influence of other psychiatric diagnoses in relatives, the hierarchical linear modeling analyses were also repeated, including only medically and psychiatrically healthy first-degree relatives (28) . Although the groups did not differ in age and parental education, because of the importance of these demographic factors the hierarchical linear modeling analyses added age, education, and parental education as covariates in the mixed model.
Estimation of Heritability
Standard maximum likelihood variance component methods implemented in SOLAR (31) were used to model estimated heritabilities. We compared a matrix of observed covariances among family members with matrices that predicted what this sharing should look like based on shared deoxyribonucleic acid (DNA), and we used maximum likelihood methods to estimate what portion of the population variance in the trait would be the result of additive genetic sharing. Thus, each individual’s performance on the neurocognitive domains was modeled as a function of measured covariates, specifically age and sex, additive genetic effects estimated from correlations among family members, and individual-specific residual environmental factors. A likelihood ratio test was used to assess statistical significance. Variance component methods generally assume that traits are normally distributed and are particularly sensitive to kurtosis in the trait distribution (32) . Use of a multivariate t distribution instead of the multivariate normal has been shown to be robust to kurtosis in the trait distribution (33) . All analyses reported in this study used the multivariate t distribution. Since 75% of participants had identical values for sensorimotor dexterity accuracy, this trait was dichotomized and analyzed using a liability threshold model (34) .
It is noteworthy that the heritability estimates calculated in multigenerational extended families are unlikely to be substantially inflated by shared environment. It would be extremely unlikely for environmental sharing to decay in a Mendelian-like manner. For shared environment to mimic genetics in an extended family, aunts, uncles, nieces, and nephews would need to be half as correlated for environment as parents, siblings, and children, and cousins would need to be half as correlated as aunts, uncles, etc., based on their expected DNA sharing. Each step on the family tree would require a fixed proportional decrease in shared environment for it to mimic the additive genetic component we are estimating and thereby inflate our heritabilities. Here lies the major advantage of the multigenerational design. When the cohort only includes nuclear families (with parents and nontwin children), it is impossible to separate out shared environment from certain types of genetic effects. The matrix that predicts additive genetic covariance structures our estimation of the presumptive heritability.
Results
Neurocognitive Profile
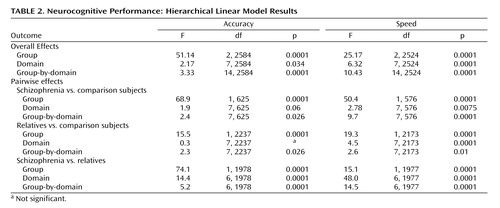
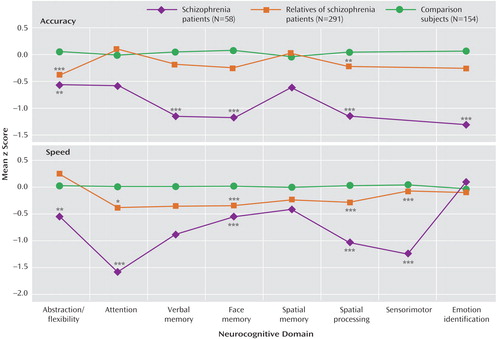
Multivariate hierarchical linear modeling effects are presented in Table 2 . For each of the two indices (accuracy, speed), hierarchical linear modeling was highly significant for group and group-by-domain interactions, warranting follow-up pairwise multivariate hierarchical linear modeling to determine the source of the significant effects. Pairwise group effects were significant, indicating that all groups differed from each other in accuracy and speed. The group-by-domain interactions were likewise highly significant for all pairwise comparisons. Group profiles, showing patients with schizophrenia, relatives, and comparison subjects, are presented in Figure 1 . As can be seen, individuals with schizophrenia performed most poorly across domains and measures, while relatives performed at an intermediate level between patients and healthy comparison subjects.
a Significance results are of univariate hierarchical linear modeling comparisons following significant overall hierarchical linear modeling analyses.
*p<0.05 versus comparison subjects; **p<0.01 versus comparison subjects; ***p<0.001 versus comparison subjects.
Although relatives performed worse than comparison subjects across neurocognitive domains, the group-by-domain interaction indicated differential effects of presumptive genetic liability on domain of impairment. Furthermore, some deficits were more pronounced for accuracy while others for speed. Most conspicuously, relatives had impaired accuracy but normal speed for abstraction and flexibility, while for attention they had normal accuracy but substantially reduced speed. There were no significant family-by-diagnosis interactions on the summary measures of the neurocognitive domains.
To evaluate the effects of genetic relatedness and add to comparability with nuclear family studies, we examined first-degree relatives separately. This group performed more poorly than comparison subjects across domains and showed a group-by-domain interaction for accuracy (group: F=31.09, df=1, 864, p<0.0001; group-by-domain: F=2.23, df=7, 864, p=0.0303) and speed (group: F=37.09, df=1, 808, p<0.0001; domain: F=2.40, df=7, 808, p=0.0195; group-by-domain: F=3.45, df=7, 808, p=0.0012). Indeed, univariate contrasts showed poorer performance in first-degree relatives for all domains except abstraction and mental flexibility, where they had decreased accuracy (p=0.0090) but normal speed (p=0.5406); attention, where they had normal accuracy (p=0.4311) but were significantly slower (p=0.0150); and spatial memory, where they did not differ from comparison subjects either in accuracy (p=0.3983) or speed (p=0.3908). On the emotion processing task they were impaired in accuracy (p<0.0001) but not speed (p=0.1585). Finally, including only psychiatrically healthy first-degree relatives yielded nearly identical results to the analysis that included relatives with other axis I disorders. Indeed, none of the differences between healthy relatives and those with another axis I disorder approached significance.
Presumptive Heritability Estimates
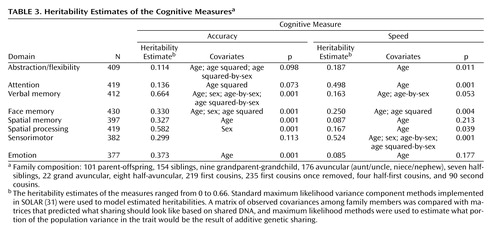
The families involved in this study provide a large number and variety of relative pairs from which the heritabilities were estimated ( Table 3 ). There were over 150 sibling pairs and over 175 cousin pairs, with relatives as distant as second cousins. However, it should be noted that the unit of analysis was actually the correlation matrix for an entire family rather than separate relative pairs.
Most measures showed age effects, with mean values declining with increasing age. Sex differences were observed on three neurocognitive domains, with women performing more accurately on verbal and face memory and men performing more accurately on the spatial task. The heritability estimates of the measures ranged from 0 to 0.69. Four neurocognitive domains (verbal memory, face memory, spatial memory, spatial processing) and emotion processing showed significant heritability estimates for accuracy and the other two (abstraction and mental flexibility, attention) showed significant heritability estimates for speed.
Discussion
We found in a multiplex multigenerational cohort that probands with schizophrenia were impaired across a range of neurocognitive domains and that relatives without schizophrenia also showed impairment in specific domains compared with healthy comparison subjects without family history. This finding confirms, with a computerized battery, earlier reports based primarily on paper and pencil tests (2 – 8) . The computerized procedure enables effective and errorless measurement of neurocognitive functions in large-scale studies, guaranteeing uniformity of data collection and scoring across sites. The finding also supports the potential of neurocognitive measures as endophenotypic markers of vulnerability to schizophrenia. Additionally, we established presumptive heritability estimates for these measures and found some to be significant, ranging from small to substantial.
The computerized measures permit additional insight into the interplay between accuracy and speed, reflecting cognitive strategies (35 , 36) . Probands and relatives were impaired overall across functions in accuracy, speed, or both. However, relatives showed considerable variability in speed for the different domains, as reflected in the group-by-domain interaction; for example, relatives had impaired performance accuracy on the abstraction and mental flexibility domain while working at normal speed. On the attention domain, by contrast, they had normal accuracy but at the expense of slowed response time. For other functions where both speed and accuracy were impaired, the impairment in speed seemed more pronounced.
This suggests that reduced speed could be a compensatory strategy that helps performance but is insufficient when the genetic vulnerability is more severe. Consequently, heritability estimates of most domains were higher for accuracy than for speed. For attention, however, where the compensatory slowing had normalized accuracy, the heritability estimate was not significant for accuracy but high for speed. Thus, examining accuracy and speed separately as endophenotypic markers should improve the specificity of detecting and interpreting genetic effects.
It is noteworthy that emotion processing, which was not examined as an endophenotypic measure in earlier studies, showed impaired accuracy in probands with intermediate accuracy in relatives, whereas both groups were less and about equally impaired for speed. The finding of impaired emotion processing abilities in relatives may relate to poorer social adaptation, which has been observed in families of patients with schizophrenia. It may also relate to the prevalence of schizotypal features observed in relatives, which include social withdrawal and awkwardness (37 , 38) .
A concern in the use of neurocognitive measures as endophenotypic markers is their susceptibility to age effect and the existence of sex differences. The present analysis incorporated an evaluation of these effects and their removal using covariance analysis. The effects we observed were consistent with the literature and buttress the sensitivity of the measures. Yet, our results also indicate that genetic variability can be established after accounting for the moderating effects of age and sex.
The study has several limitations. The unique multiplex multigenerational cohort may yield results that could differ from studies of simplex families with first-degree relatives. However, the neurocognitive profile obtained in the present cohort is similar to that obtained in sporadic schizophrenia (19) and in other familial cohorts. Furthermore, in a multigenerational design heritability estimates are less likely to be inflated by effects of shared environment, as is the case in studies of first-degree relatives only. Notably, while heritability estimates of most domains were significant, their magnitude was not as high as reported in some twin studies for DSM-based diagnosis. Heritability and familial environment can be confounded in studies of nuclear families, but the present analysis is somewhat protected from this influence because for heritability estimates to be inflated by shared environment, the degree of environmental sharing would have to drop off with the degree of relationship in a manner that resembles Mendelian laws.
In choosing endophenotypes for genetic studies, we need measures that are associated with disease, that differentiate at-risk individuals, and that are heritable. The present results indicate that several neurocognitive measures fulfill these criteria. Specifically, memory and emotion processing accuracy and speed of attention have moderate to strong genetic influences on variation in performance levels between individuals. These traits would be sensible targets for genome scans to identify loci influencing variation in these disease-related risk factors (39 , 40) .
1. Harrison PJ, Weinberger DR: Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry 2005; 10:40–68Google Scholar
2. Saykin AJ, Gur RC, Gur RE, Mozley PD, Mozley LH, Resnick SM, Kester DB, Stafiiak P: Neuropsychological function in schizophrenia: selective impairment in memory and learning. Arch Gen Psychiatry 1991; 48:618–624Google Scholar
3. Elvevag B, Goldberg TE: Cognitive impairment in schizophrenia is the core of the disorder. Crit Rev Neurobiol 2000; 14:1–21Google Scholar
4. Cannon TD, Zorrilla LE, Shtasel DL, Gur RE, Gur RC, Marco EJ, Moberg PJ, Price RA: Neuropsychological functioning in siblings discordant for schizophrenia and healthy volunteers. Arch Gen Psychiatry 1994; 51:651–661Google Scholar
5. Faraone SV, Seidman LJ, Kremen WS, Pepple JR, Lyons MJ, Tsuang MT: Neuropsychological functioning among the non-psychotic relatives of schizophrenic patients: a diagnostic efficiency analysis. J Abnorm Psychol 1995; 104:286–304Google Scholar
6. Egan MF, Goldberg TE, Gscheidle T, Weirich M, Rawlings R, Hyde TM, Bigelow L, Weinberger DR: Relative risk for cognitive impairments in siblings of patients with schizophrenia. Biol Psychiatry 2001; 50:98–107Google Scholar
7. Thompson JL, Watson JR, Steinhauer SR, Goldstein G, Pogue-Geile MF: Indicators of genetic liability to schizophrenia: a sibling study of neuropsychological performance. Schizophr Bull 2005; 31:85–96Google Scholar
8. Conklin HM, Curtis CE, Calkins ME, Iacono WG: Working memory functioning in schizophrenia patients and their first-degree relatives: cognitive functioning shedding light on etiology. Neuropsychologia 2005; 43:930–942Google Scholar
9. Gottesman II, Gould TD: The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry 2003; 160:636–645Google Scholar
10. Freedman R, Adler LE, Leonard S: Alternative phenotypes for the complex genetics of schizophrenia. Biol Psychiatry 1999; 45:551–558Google Scholar
11. Williams JT, Blangero J: Comparison of variance components and sibpair-based approaches to quantitative trait linkage analysis in unselected samples. Genet Epidemiol 1999; 16:113–134Google Scholar
12. Shedlack K, Lee G, Sakuma M, Xie SH, Kushner M, Pepple J, Finer DL, Hoff AL, DeLisi LE: Language processing and memory in ill and well siblings from multiplex families affected with schizophrenia. Schizophr Res 1997; 25:43–52Google Scholar
13. Faraone SV, Seidman LJ, Kremen WS, Toomey R, Pepple JR, Tsuang MT: Neuropsychologic functioning among the nonpsychotic relatives of schizophrenic patients: the effect of genetic loading. Biol Psychiatry 2000; 48:120–126Google Scholar
14. Tuulio-Henriksson A, Arajarvi R, Partonen T, Haukka J, Varilo T, Schreck M, Cannon T, Lonnqvist J: Familial loading associates with impairment in visual span among healthy siblings of schizophrenia patients. Biol Psychiatry 2003; 54:623–628Google Scholar
15. Hoff AL, Svetina C, Maurizio AM, Crow TJ, Spokes K, DeLisi LE: Familial cognitive deficits in schizophrenia. Am J Med Genet B Neuropsychiatr Genet 2005; 133:43–49Google Scholar
16. Nurnberger JI, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T/NIMH Genetics Initiative: Diagnostic Interview for Genetic Studies: rationale, unique features, and training. Arch Gen Psychiatry 1994; 51:849–859Google Scholar
17. Maxwell ME: Manual for the Family Interview for Genetic Studies (FIGS). Bethesda, Md, Clinical Neurogenetics Branch, Intramural Research Program, National Institute of Mental Health, 1992Google Scholar
18. Gur RC, Ragland JD, Moberg PJ, Turner TH, Bilker WB, Kohler C, Siegel SJ, Gur RE: Computerized neurocognitive scanning, I. methodology and validation in healthy people. Neuropsychopharmacology 2001; 25:766–776Google Scholar
19. Gur RC, Ragland JD, Moberg PJ, Bilker WB, Kohler C, Siegel SJ, Gur RE: Computerized neurocognitive scanning, II. the profile of schizophrenia. Neuropsychopharmacology 2001; 25:777–788Google Scholar
20. Kurtz MM, Ragland JD, Moberg PJ, Gur RC: The Penn Conditional Exclusion Test: a new measure of executive-function with alternate forms of repeat administration. Arch Clin Neuropsychol 2004; 19:191–201Google Scholar
21. Kurtz MM, Ragland JD, Bilker W, Gur RC, Gur RE: Comparison of the Continuous Performance Test with and without working memory demands in healthy controls and patients with schizophrenia. Schizophr Res 2001; 48:307–316Google Scholar
22. Gur RC, Jaggi JL, Ragland JD, Resnick SM, Shtasel D, Muenz L, Gur RE: Effects of memory processing on regional brain activation: cerebral blood flow in normal subjects. Int J Neurosci 1993; 72:31–44Google Scholar
23. Glahn DC, Gur RC, Ragland JD, Censits DM, Gur RE: Reliability, performance characteristics, construct validity, and an initial clinical application of a Visual Object Learning Test (VOLT). Neuropsychology 1997; 11:602–612Google Scholar
24. Benton AL, Varney NR, Hamsher KS: Judgment of Line Orientation, Form V. Iowa City, Iowa, University of Iowa Hospitals, 1975Google Scholar
25. Gur RE, Kohler CG, Ragland JD, Siegel SJ, Lesko K, Bilker WB, Gur RC: Flat affect in schizophrenia: relation to emotion processing and neurocognitive measures. Schizophr Bull 2006; 32:279–287Google Scholar
26. Bryk AS, Raudenbush SW: Hierarchical Linear Models. Newbury Park, Calif, Sage Publications, 1992Google Scholar
27. Singer J: Using SAS PROC MIXED to fit multilevel models, hierarchical models, and individual growth models. J Edu Behav Stat 1998; 24:323–355Google Scholar
28. Calkins ME, Curtis CE, Iacono WG: Antisaccade performance is impaired in medically and psychiatrically healthy biological relatives of schizophrenia patients. Schizophr Res 2004; 71:167–178Google Scholar
29. Calkins ME, Gur RC, Ragland JD, Gur RE: Face recognition memory deficits in schizophrenia patients and their relatives: comparison with visual object memory performance. Am J Psychiatry 2005; 162:1963–1966Google Scholar
30. Rabe-Hesketh S, Toulopoulou T, Murray RM: Multilevel modeling of cognitive function in schizophrenic patients and their first degree relatives. Multivariate Behav Res 2001; 36:279–298Google Scholar
31. Almasy L, Blangero J: Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 1998; 62:1198–1211Google Scholar
32. Allison DB, Neale MC, Zannolli R, Schork NJ, Amos CI, Blangero J: Testing the robustness of the likelihood-ratio test in a variance-component quantitative-trait loci-mapping procedure. Am J Hum Genet 1999; 65:531–544Google Scholar
33. Blangero J, Williams JT, Almasy L: Robust LOD scores for variance component based linkage analysis. Genet Epidemiol 2000; 19(Suppl):S8–S14Google Scholar
34. Williams JT, Van Eerdewegh P, Almasy L, Blangero J: Joint multipoint linkage analysis of multivariate qualitative and quantitative traits, I: likelihood formulation and simulation results. Am J Hum Genet 1999; 65:1134–1147Google Scholar
35. Meyer DE, Irwin DE, Osman AM, Kounios J: The dynamics of cognition and action: mental processes inferred from speed-accuracy decomposition. Psychol Rev 1988; 95:183–237Google Scholar
36. Smith RW, Kounios J: Sudden insight: all-or-none processing revealed by speed-accuracy decomposition. J Exp Psychol Learn Mem Cogn 1996; 22:1443–1462Google Scholar
37. Nuechterlein KH, Asarnow RF, Subotnik KL, Fogelson DL, Payne DL, Kendler KS, Neale MC, Jacobson KC, Mintz J: The structure of schizotypy: relationships between neurocognitive and personality disorder features in relatives of schizophrenic patients in the UCLA Family Study. Schizophr Res 2002; 54:121–130Google Scholar
38. Calkins ME, Curtis CE, Grove WM, Iacono WG: Multiple dimensions of schizotypy in first degree biological relatives of schizophrenia patients. Schizophr Bull 2004; 30:317–325Google Scholar
39. Fanous AH, Kendler KS: Genetic heterogeneity, modifier genes, and quantitative phenotypes in psychiatric illness: searching for a framework. Mol Psychiatry 2005; 10:6–13Google Scholar
40. Hallmayer JF, Kalaydjieva L, Badcock J, Dragovic M, Howell S, Michie PT, Rock D, Vile D, Williams R, Corder EH, Hollingsworth K, Jablensky A: Genetic evidence for a distinct subtype of schizophrenia characterized by pervasive cognitive deficit. Am J Hum Genet 2005; 77:468–476Google Scholar